Several studies have reported an association between tumor necrosis factor α (TNF-α) polymorphisms and inflammatory diseases such as psoriasis vulgaris and psoriatic arthritis, although the results vary according to the population studied. No studies have been performed in the Spanish population.
ObjectiveTo analyze the polymorphisms of the promoter region of the TNF-α gene in patients with moderate to severe psorasis and to identify potential differences in genotype compared to a group of healthy volunteers.
Material and methodsEighty-nine patients with moderate to severe psoriasis and 76 healthy controls with no personal or family history of psoriasis were selected. Polymorphisms of the TNF-α promoter region of both groups were genotyped.
ResultsWe observed a higher prevalence of the genotype with both wild-type alleles at positions -238 (GG genotype, 86.5% vs 70.4%, respectively) and -1031 (TT genotype, 80.2% vs 45.8%, respectively) in patients compared to the healthy control group. The differences at positions -308 and -857 were not significant.
ConclusionThere are differences in polymorphisms at positions -238 and -1031 in patients with moderate to severe psoriasis compared to healthy volunteers. This observation provides further support for the importance of the part that TNF-α plays in the pathophysiology of this disease.
En diversos estudios se ha descrito la asociación de los polimorfismos del factor de necrosis tumoral α (TNF-α) y patologías inflamatorias como la psoriasis vulgar y la artritis psoriásica, aunque los resultados son variables según la población de origen de la muestra. No se han realizado estudios previamente en población española.
ObjetivoAnalizar los polimorfismos de la región promotora del gen del TNF-α en pacientes con psoriasis moderada-grave y establecer las posibles diferencias genotípicas con los hallados en un grupo de voluntarios sanos.
Material y métodosSe seleccionaron 89 pacientes con psoriasis moderada-grave y 76 controles sin antecedentes familiares ni personales de psoriasis. Se genotiparon los polimorfismos de la región promotora del gen del TNF-α en ambos grupos.
ResultadosObservamos una mayor prevalencia de genotipo con ambos alelos en estado silvestre en posición -238 (GG, 86,5% vs 70,4% respectivamente) y -1031 (TT, 80,2% vs 45,8% respectivamente) en el grupo de pacientes con psoriasis al compararlo con el grupo control. Las diferencias halladas en posición -308 y -857 no fueron significativas.
ConclusiónExisten diferencias en los polimorfismos en las posiciones -238 y -1031 entre pacientes con psoriasis moderada-grave y voluntarios sanos, lo cual apoya la importancia del papel del TNF-α en la fisiopatología de esta entidad.
One of the main problems in clinical pharmacology is dealing with interindividual variability in the effects of drugs, both in terms of efficacy and safety.1 The same drug can be very effective in one patient but lead to adverse reactions that require discontinuation of treatment in another. This variability is due to both genetic factors, which account for 20% to 95% of the variability in the bioavailability and effect of the drug,2 and nongenetic factors, which unlike genetic factors can vary during an individual's lifespan. These nongenetic factors include physiological factors (age, sex, weight, and body fat), pathophysiological factors (liver, kidney, and cardiovascular function, and associated diseases), and environmental factors (tobacco and alcohol consumption and concomitant treatments).
The main challenge facing medicine today is identifying the variables that can predict response to a drug or its toxicity in each individual prior to treatment. Pharmacogenetics is the study of genetic characteristics and their relationship with pharmacological response, both in terms of efficacy and adverse reactions. The first step in this process is to study individual genetic variations.
In the case of psoriasis, studies have attempted to identify genetic polymorphisms that act as markers of disease susceptibility. The main genes analyzed to date are the major histocompatibility complex HLA-Cw*06023 and the genes encoding tumor necrosis factor (TNF) α,4–10 various interleukins such as IL-1β, IL-6, and IL-10,11,12 subunit p40 common to IL-12 and IL-23, a subunit of the IL-23 receptor,13 IL-13 and IL-15,14 SNF313 (a gene implicated in protein ubiquitination),15 and transforming growth factor β,16 as well as the promoter of the gene encoding interferon-γ.17
Of particular interest in psoriasis is the study of polymorphisms in the TNF-α gene, given that most biologics used to treat the disease target TNF-α protein.18–21TNF-α is located on the short arm of chromosome 6, very close to the major histocompatibility complex B. This region is highly polymorphic, with up to 44 polymorphisms reported,22 some of which have been associated with different diseases. Polymorphisms at positions 238 and 308 of the TNF-α promoter have been associated in some studies with response to anti-TNF-α biologics in rheumatologic diseases such as rheumatoid arthritis and ankylosing spondilitis.22–29 Polymorphisms at position 857 have also been associated with response to other biologics in patients with rheumatoid arthritis.30,31 A study in patients with rheumatoid arthritis, published by Oregón-Romero et al.32 showed that those patients with 2 wild-type alleles at both position 238 and position 308 had higher levels of TNF-α messenger RNA and TNF-α protein production than those with 1 mutant allele. The same differences were not found in healthy individuals, however.
In psoriasis, the most widely studied polymorphisms are substitution of guanine by adenine at positions 238 and 308 (-238G→A, -308G→A), substitution of cytosine by thymine at position 857 (-857C→T), and substitution of thymine by cytosine at position 1031 (-1031T→C). The first 2 polymorphisms, located in the promoter region of the gene, have been associated with the severity of psoriasis,32 whereas the presence of -857T has been associated with a greater risk of psoriatic arthritis, but not psoriasis. However, no conclusive associations have been found for polymorphisms at position 1031.9
ObjectiveThe primary objective of this study was to investigate genetic polymorphisms in the TNF-α promoter in patients with psoriasis and control subjects and to establish possible allelic and genetic differences between the 2 groups.
Materials and MethodsStudy DesignThis was a prospective study in patients with moderate to severe plaque psoriasis, defined as a psoriasis area and severity index greater than or equal to 10 and/or an affected body surface area greater than 10%, with or without psoriatic arthritis. All patients were resident in the autonomous communities of Madrid or Castile-La Mancha, Spain. Patients did not have to be of Spanish origin to be included, although 97% of them were.
For the control group, we used healthy volunteers from the clinical trials unit of the clinical pharmacology department. To be included, the healthy controls had to be residents of the autonomous community of Madrid and be nonsmokers of Spanish descent with no personal or family history of psoriasis in first-degree relatives.
The study was approved by the ethics committee of our hospital. Informed consent was obtained from all healthy volunteers prior to sample collection.
For this study, we selected the following most widely studied polymorphisms in patients with psoriasis: -238 G→A (rs361525), -308 G→A (rs1800629), -857 C→T (rs1799724), and -1031 T→C (rs1799964).
Sample ProcessingFor each participant, a 4-mL sample of peripheral blood was extracted to a K3 ethylenediaminetetraacetic acid tube, which was duly labeled with an identifying code. DNA was extracted using an automated system (MagNa Pure® System, Roche Applied Science, USA) and quantified by spectrophotometry (NanoDrop® ND-1000, Wilmington DE, USA). Sample purity was determined by measuring the 260nm/280nm signal ratio.
Polymerase Chain ReactionSingle nucleotide polymorphisms (SNP) were determined by sequencing with different primers. Primers 308F (5′-TTCCTGCATCCTGTCTGGAA-3′) and 238R (5′-CAGCGGAAAACTTCCTTGG-3′) were used for polymorphisms -308 and -238, respectively. These polymorphisms were studied in the same sequence given their proximity to one another on the gene. Primers 857F (5′-AGGAATGGGTTACAGGAGAC-3′) and 857R (GTCCCCTGTATTCCATACCT) were used for polymorphism -857, whereas primers 1031F (5′-TCAGAGAGCTTCAGGGATAT-3′) and 1031R (5′-ACATGTGGCCATATCTCCCA-3′) were used for polymorphism -1031.
The reagent concentrations in the polymerase chain reaction (PCR) were 1X buffer, 2.5mM MgCl2, 0.3μM deoxyribonucleotide triphosphate (dNTP), and 1 U/μL Ampli Taq Gold© (Applied Biosystems). In addition, for study of the nucleotide at position -1031, we added 5% dimethyl sulfoxide, as this is a region rich in the nitrogenous bases guanine and cytosine.
The PCR product was resolved on a 3% agarose gel and purified with GENECLEAN© (MP Biomedicals), according to the manufacturer's instructions. The amplified product was then sequenced.
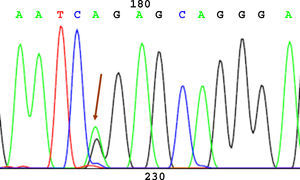
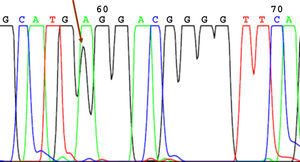
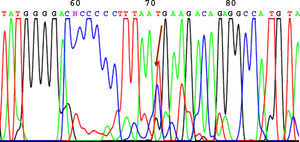
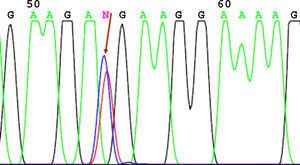
SequencingBefore sequencing, PCR was performed with BigDye® Terminator v3.1 (Applied Biosystems), with the first primer applied directly. Following this PCR, the product was purified once again to remove remnants of fluorescently labeled dNTP. Fluorescence was detected by the sequencer (Applied Biosystems). The sequencing results were analyzed using the Chromas V 1.45 software. Figures 1 to 4 show examples of sequences for polymorphisms at positions -238, -308, -857, and -1031.
Statistical AnalysisThe SPSS program, version 15.0.1, was used for statistical analysis. For quantitative variables, the Pearson χ2 test was applied to analyze the differences in genotype frequencies between patients with moderate to severe psoriasis and the group of healthy volunteers for each of the 4 polymorphisms studied.
ResultsWe analyzed the DNA from 89 patients and 76 healthy individuals. The epidemiological characteristics of the participants are shown in Table 1. Sequencing of the 4 polymorphisms was performed in 70 cases and 69 controls.
Characteristics of Cases and Controls.
| Controls (n=76) | Cases (n=89) | |
| Mean age, y (range) | 22.60 (18-31) | 48.17 (19-80) |
| Sex | ||
| Male, % | 46% | 51.7% |
| Female, % | 54% | 48.3% |
| Male/female ratio | 1.2/1 | 0.9/1 |
| Inclusion criteria | Healthy volunteers | Moderate to severe psoriasis |
| Lack of family and personal history of psoriasis | ||
| Nonsmokers | ||
The distribution of allele and genotype frequencies analyzed so far are shown in Table 2. Sequencing of polymorphism -238 showed a greater frequency of wild-type allele G, as well as a greater frequency of the genotype with both wild-type alleles (GG) in patients with psoriasis than in the group of healthy volunteers (86.5% vs 70.4%). This difference was statistically significant (P=.050).
Comparison of Allele and Genotype Frequencies Between Cases and Controls.
| SNP (No. Patients) | Cases | Controls | P Value for Statistical Significance (Genotype Frequencies) | ||
| Allele Frequencies | Genotype Frequencies | Allele Frequencies | Genotype Frequencies | ||
| -238 G→A | G 92.6%A 7.4% | G/G 70/81 (86.5%)A/G 10/81 (12.3%)A/A 1/81 (1.2%) | G 83.1%A 16.9% | G/G 50/71 (70.4%)A/G 18/71 (25.4%)A/A 3/71 (4.2%) | P=.050 |
| -308 G→A | G 91.7%A 8.3% | G/G 70/84 (83.3%)A/G 14/84 (16.7%)A/A 0 | G 89.5%A 10.5% | G/G 60/76 (78.9%)A/G 16/76 (21.1%)A/A 0 | P=.478 |
| -857 C→T | C 87%T 13% | C/C 58/77 (75.3%)C/T 18/77 (23.4%)T/T 1/77 (1.3%) | C 92.1%T 7.9% | C/C 65/76 (85.5%)C/T 10/76 (13.2%)T/T 1/76 (1.3%) | P=.262 |
| -1031 T→C | T 80.2%C 19.8% | T/T 55/86 (69.4%)T/C 28/86 (28.6%)C/C 3/86 (2.0%) | T 66.7%C 33.3% | T/T 33/72 (45.8%)T/C 30/72 (41.7%)C/C 9/72 (12.5%) | P=.025 |
Abbreviation: SNP, single nucleotide polymorphism.
We observed a higher frequency of -308GG in patients than in controls (83.3% vs 78.9%), although this difference was not statistically significant (P=.478). At position -857, the presence of a genotype with both wild-type alleles (CC) was more frequent in controls (85.5%) than in patients (75.3%), although this difference was also not significant (P=.262).
For sequencing of polymorphism -1031, we found a higher frequency of the wild-type T allele in patients with psoriasis (80.2% vs 66.7%); 69.4% of the patients had a genotype with both wild-type alleles (TT), whereas this genotype was only present in 45.8% of controls. This difference was statistically significant (P=.025).
DiscussionTNF-α is a proinflammatory cytokine implicated in the pathophysiology of psoriasis and other inflammatory diseases such as rheumatoid arthritis and Crohn disease. Several polymorphisms have been associated with psoriasis.
We found an association between some polymorphisms of TNF-α and susceptibility to developing psoriasis. We also found differences in the genetic sequences of the promoter-region polymorphisms TNFα -238 and TNFα -1031 on comparing a population with moderate to severe psoriasis with a population of healthy volunteers. This leads us to think that these polymorphic differences might play an important role in the pathophysiology of psoriasis.
The greater frequency of -238GG and -308GG in patients with psoriasis than in controls could be explained by the greater production of TNF-α in these subjects, as demonstrated in messenger RNA studies performed in other diseases in which TNF-α levels are increased.32 The greater frequency of -1031TT in patients with psoriasis could also be explained the same way, although few studies of the frequency of this polymorphism in patients with psoriasis have been undertaken and those that have did not find any differences; in addition, there are no relevant in vitro data for this polymorphism.
We observed that the wild-type allele TNFα -238G occurred more frequently in patients with psoriasis (92.6%) than in the control group (83.1%). The GG genotype also occurred more frequently (86.5% vs 70.4%). Most studies published by other authors have found a greater proportion of mutant TNFα -238A allele among patients with psoriasis, as well as an increased risk of psoriasis in association with genotypes with some of the mutant alleles (GA/AA).4–6,9,33 However, a recently published study concluded that the -238GG genotype is more frequent among individuals with more severe forms of psoriasis,34 as was the case in our study. Some studies have found these differences only in patients with early-onset psoriasis,5,6 or only in male patients.6 Studies performed in an oriental population did not find any differences in the distribution of these polymorphisms.7,35,36
We did not find any significant differences in the distribution of the TNFα -308 genotype, in line with the findings of other authors.4,7,9,10,35,36 However, some studies have found a higher frequency of TNFα -308GG in the case of moderate to severe psoriasis,37 or a higher frequency of the TNFα -308G allele in patients with early-onset psoriasis.5,8,33,38 In a meta-analysis published by Li et al.39 in 2007, which included studies published to date on polymorphisms of TNF-α at position -238 and -308, the authors concluded that the presence of a wild-type G allele might play a protective role in psoriasis.
Likewise, TNF-α polymorphisms at position -857 did not show any differences in distribution in our study. Similar studies performed in other populations have not found differences in psoriasis,9 although a higher frequency of the mutant TNFα -857 allele has been observed in patients with psoriatic arthritis compared to healthy volunteers.9,40
We found differences in the genotype distribution of TNFα -1031 between patients and controls, with a greater frequency of the wild-type TT genotype among patients. To the best of our knowledge, the only group to have studied this polymorphism did not find statistically significant differences.9
We should bear in mind that these differences in the polymorphism of the TNF-α gene in published studies could be due to differences in population or race.41
Our study is subject to certain limitations. The most important is the small sample size both for patients and controls. In addition, there was a substantial difference in age (and a less marked difference in sex) between cases and controls. However, we do not consider these imbalances to be important because the genotype remains almost completely stable throughout an individual's life. In addition, the fact that we only assessed those patients with moderate to severe psoriasis and that we did not adjust for whether psoriatic arthritis was present may have introduced a bias given the findings of published studies. Finally, patients and controls were drawn from the Communities of Madrid and Castile-La Mancha. The same study with samples from residents of other regions would be needed to determine whether these results can be extrapolated to the Spanish population as a whole.
To perform this study, a gene database was created for the study of the TNF-α polymorphisms. This is very important as, to date, we are unaware of databases for polymorphisms of the TNF-α receptor gene in Spain in psoriasis patients and controls. Moreover, the literature on the subject is limited. Genotyping of these polymorphisms of TNF-α is the first step in the future study of new polymorphisms, both of TNF-α and protein p40, and also of other proteins implicated in the pathophysiology and treatment of psoriasis. In addition, this genetic analysis associated with the assessment of the safety and efficacy of study drugs could allow us to study possible genetic differences between patients according to therapeutic response. The main aim of pharmacogenetics is to establish genetic differences between individuals, and so enable optimization of the treatment of diseases in each patient, thereby ensuring the patient receives safer therapy. This would allow us to avoid delays in the administration of an effective therapy and avoid risks for the patient. It would also prevent unnecessary spending on ineffective treatments.
We should be aware that the genetic profile not only has an influence on the development of a given illness, but also the way in which other genes are expressed.41 Therefore, it would be interesting to perform future analyses of gene expression by techniques such as DNA microarrays, thereby providing not only a qualitative analysis but also a quantitative one.42
The aim of this study has thus been to take a further step forward in the field of pharmacogenetics.
ConclusionsIn our study, we would like to highlight that:
- 1.
We found significant differences in polymorphism -1031 in psoriasis patients compared to the control group, suggesting that this mutation could play an active part in the pathophysiology of this disease. This finding has not been reported previously in any genetic study of patients with moderate to severe psoriasis.
- 2.
We also found relevant differences in polymorphism -238, and so this mutation may also participate in the genesis of this disease. This finding has not been reported previously in Spanish patients with moderate to severe psoriasis.
- 3.
Detection of these polymorphisms is of notable practical worth as, for present or future studies, it would enable:
- a.
creation of a gene bank with blood samples obtained from patients with plaque psoriasis;
- b.
study of the pathogenic mechanisms underlying psoriasis;
- c.
identification of other possible therapeutic targets;
- d.
correlation of these results with treatment response and toxicity, whereby we would achieve:
- -
an increase in the knowledge of the mechanism of action at the molecular level of the study drugs and
- -
determination a priori of the drug of choice (personalized medicine).
- -
- a.
This study received financial support from the Fundación Salud 2000 and was partially funded by the FIS PI10-01740 project. The study was awarded the Juan de Azúa prize in 2010 by the Spanish Academy for Dermatology and Venereology.
Conflicts of InterestThe authors declare that they have no conflicts of interest.
Please cite this article as. Gallo E, et al. Estudio de los polimorfismos genéticos de la región promotora del TNF-α en pacientes españoles afectos de psoriasis. Actas Dermosifiliogr.2012;103:301-7.