The emergence of new technologies and the management of huge amounts of data have revolutionized all areas of knowledge. This effect is increasingly evident in medicine, economics, education, humanities, and other spheres of our societies.
The dermatologist’s clinical practice one area affected by this inevitable change.1 The application of new targeted therapies has lead to changes in our treatment protocols for some diseases, including psoriasis, atopic dermatitis, and advanced melanoma. Thanks to these advances, we have positioned ourselves at the forefront of immunology, gene therapy, nanotechnology, and telemedicine.2
In parallel, technological advances have led to an explosion in the generation of digital data and improvements in algorithms and computer hardware.3 In the technological age, innovation entails the development not only of new drugs, but also of new techniques and diagnostic methods that can improve patient care.1
The use of mathematical models to seek answers to medical and biological questions is now a reality. The recent discovery of a new geometric form, the scutoid, has helped explain the architecture and packing of curved epithelia, facilitating greater understanding of the 3-dimensional organization of epithelial organs.4 In the field of dermatology, fractal geometry has been used as a method for the evaluation of melanocytic lesions. The analysis of lacunarity, initially used to characterize irregular geometric objects whose structures contain repeating units of similar structure at various scales of magnification (fractals), has enabled distinction of nevi and melanomas with a sensitivity and specificity of 92% and 81%, respectively.5
In the emerging era of globalized knowledge, everything that can be automated or calculated probably will be. In recent years, medicine has been standardized and regulated to facilitate the diagnosis and treatment of a large number of diseases. However, the use of algorithms in clinical practice also enables the application of artificial intelligence to medical decision-making processes.1
So-called machine learning is a form of artificial intelligence that seeks to develop techniques that allow computers to learn. In general, machine learning refers to the wide range of algorithms that make intelligent predictions based on the automated analysis of large datasets.3,6 Machine learning is therefore a broad concept that ranges from simple decision trees or linear regression to complex artificial neural networks involving millions of data points.3
The emergence of a scientific method based on extrapolation of data through random sampling was a consequence of our inability to analyze and interpret all available data.7 Deep learning has emerged as a computational tool to respond to this unmet need.
Deep learning is based on the use of artificial neural networks designed after the cells of the cerebral cortex.8 This framework allows the representation of high-level data, and the creation of millions of parameters for segmentation, classification, and image detection.3,9 Specifically, convolutional neural networks (CNNs) and multilayer perceptrons have revolutionized the field of pattern detection in medical imaging.3,6 These networks combine multiple, specialized, hierarchical layers of learning with sophisticated filters to form deep neural networks capable of detecting complex shapes.
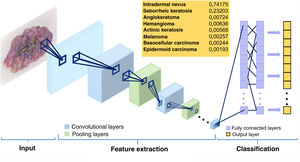
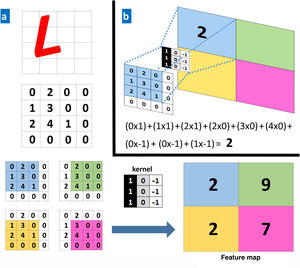
The convolutional layers receive an input, which they transform and transmit to the next layer. This modification (convolution operation) is performed via filters or detectors of established patterns (kernels) that produce a new, reduced and increasingly abstract representation of the input (feature maps). Pooling layers aggregate similar data by taking the mean, maximum, or other statistics of the feature maps, thus further reducing the variability of the data. The final (fully connected) layers assign the data a particular label after performing normalization and rectification operations. In this way, the input image is modeled through successive convolutional and nonlinear operations until it is converted to a probability distribution of belonging to a particular class of image9 (Figs. 1 and 2).
Schematic illustrating the operation of a convolutional neural network. The input image is transformed through the successive layers (convolution and pooling layers). The final (fully connected) layers classify the data to reach a given diagnosis.11
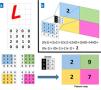
A, Example of a convolution operation. A computer sees an image (L) as a 4×4 matrix of numbers. Each value of the input image is multiplied by a filter or kernel. Consequently, a new, reduced representation is obtained (a 2×2 feature map). B, Example of a specific operation for the values in the blue box.
Deep learning is progressing towards its ultimate goal: artificial intelligence.8 Most of the prediction models used in medical imaging are trained using collections of prelabeled examples (supervised learning).3,8 The great advances in information technology of recent years allow neural networks to assimilate large amounts of data, increasing the number of connections between the different layers (neurons), which can then work cooperatively to solve increasingly complex cases.9 Furthermore, the great advances of neural networks are based on the fact that, unlike traditional artificial intelligence, they can autonomously create rules that allow them to classify a certain characteristic based on raw data.3,9–11 Thus, these networks can learn by themselves, optimizing their predictions and improving their efficacy.
Thanks to these abilities, the application of CNNs to medical imaging holds great promise. Specialties such as dermatology, radiology, and pathological anatomy are ideal fields for the application of systems based on classification, segmentation, and image detection.3
In 2017, Esteva et al6 evaluated the performance of CNNs for the clinical evaluation of skin lesions. Using 127463 images of lesions of epidermal and melanocytic origin, the authors compared the diagnostic capacity of 21 expert dermatologists with that of a trained CNN on clinical images with 2 critical binary classification use cases (benign or malignant). The specialists correctly identified 95% of the malignant tumors and 76% of the benign lesions, while the CNN correctly diagnosed 96% and 90% of cases, respectively. In another study, an automated learning fusion algorithm performed better than most dermatologists in classifying 100 dermoscopic images of nevi and melanomas.12
The dermatological applications of neural networks are not limited to skin cancer. DeepGestalt is a system that combines visual recognition and artificial intelligence technologies to identify facial phenotypes of genetic disorders. This program outperformed clinicians in detecting syndromes, and could improve the performance of genetic testing.13 Han and coworkwers14 evaluated the accuracy of a CNN trained with 49567 images for the diagnosis of onychomycosis, and demonstrated that the deep learning-based system performed better than most of the dermatologists who participated in the study.
The last year has seen an explosion in the number of similar studies. The apparently inexorable progress of these systems raises important questions: will these systems ever replace the dermatologist, and how will these changes be integrated into dermatology in the future?15,16
Therapeutic decisions could be made and diagnostic algorithms developed based on better scientific evidence. Computers, with their unlimited learning capacity, will replace many decision-making processes that are currently exclusively carried out by humans. So-called big data refers to datasets, or combinations thereof, of such volume, complexity, and rapid growth that processing and analysis using conventional technologies and tools is impossible. Analysis of these datasets will allow us to avoid the inevitable biases generated by randomness. The success of artificial intelligence will depend on the participation of dermatologists in the development of appropriate algorithms and the use of large public archives that allow the results to be generalized.17 The use of blockchain technology will extend beyond financial applications (e.g. Bitcoin), making it possible to store and exchange coded images, and offering solutions to privacy, security, and data distribution issues.18
Despite the potential benefits of artificial intelligence, many of the implications of the use of this technology remain unknown,19 and most of these systems have not been evaluated in a real clinical setting. In practice, medical decision-making requires more than morphological criteria alone.16 Patient management is a complex and multifactorial process that, in addition to clinical data, must take into account alternatives (e.g. findings obtained via strict follow-up) to exclusively binary endpoints (benign versus malignant, removal versus no treatment required).3,16 Mathematical models do not establish a diagnosis, but provide a diagnostic probability, a value of 0 or 1 that in the future could be used to guide less experienced doctors or even serve as a screening method, always complementing, but not replacing, the capabilities of the dermatologist.15,20
Algorithms will be incorporated into dermatological practice and will increase diagnostic accuracy in the hands of experts.12 However, in the context of the doctor-patient relationship, information and knowledge represent just 2 of the many elements required to practice quality medicine. Human factors will continue to play a fundamental role, which cannot be replaced by computers. Other key aspects of good medical practice, including empathy and patient support, common sense, creativity, passion, motivation, and innovation, make the figure of the dermatologist irreplaceable.
Conflicts of InterestThe authors declare that they have no conflicts of interest.
Please cite this article as: Iglesias-Puzas Á, Boixeda P. Deep learning y DerMATología. Actas Dermosifiliogr. 2020;111:192–195.