Melanoma is a malignant neoplasm with high metastatic disease risk and elevated mortality. Incidence of melanoma varies according to geographic region and genetic background. Epidemiological studies indicate that acral melanoma (AM) is among the most common melanomas in the Mexican population. While extensive studies have identified genes associated with melanoma, little is known about the genes involved in the pathogenesis of AM.
ObjectiveTo compare the gene expression patterns between primary melanoma and normal skin.
MethodsWe used 10 samples of fresh acral melanomas and normal skin for the study of differential gene expression and 22 samples of melanoma for in situ hybridization.
ResultsWe first identified a gene that was present in a sample of AM and absent in normal skin. DNA sequencing of this differentially expressed gene revealed that it corresponded to ABCB5, a gene recently implicated in the regulation of progenitor cell fusion. Furthermore, we detected ABCB5 expression in other melanoma specimens by RT-PCR. We showed that nine out of ten melanomas were positive for ABCB5 while only one melanoma and normal skin samples were negative. All ABCB5 expressing melanomas had variable gene expression according to in situ hybridization studies, suggesting that the ABCB5 gene may be differentially regulated by individual melanomas.
ConclusionsThe ABCB5 gene may be related to the properties of chemoresistance and aggressiveness of melanoma. The high expression found in samples of acral melanoma may provide more insight on the pathogenesis of this common type of melanoma in the Mexican population, frequently associated with poor prognosis.
El melanoma es una neoplasia maligna que presenta una elevada mortalidad y un alto riesgo de desarrollar metástasis. La incidencia de esta enfermedad varía en función de la región geográfica y el trasfondo genético. Estudios epidemiológicos indican que el melanoma acral es uno de los más comunes en la población mexicana. Hay una gran cantidad de estudios sobre genes asociados al melanoma, sin embargo, se sabe muy poco de los genes que se relacionan con el melanoma acral.
ObjetivoComparar el patrón de expresión génica entre melanomas acrales primarios y piel sana.
MétodosSe utilizaron muestras en fresco de 10 lesiones de melanoma acral y piel sana para el estudio de expresión diferencial de genes y 22 muestras de melanoma para la hibridación in situ.
ResultadosIdentificamos un gen que estaba presente en una muestra de melanoma acral y ausente en la piel normal. La secuenciación de este gen reveló que correspondía al gen ABCB5, recientemente implicado en la regulación de la fusión de células progenitoras. Al realizar RT-PCR de otros melanomas se detectó la expresión de este gen: 9 de 10 melanomas fueron positivos para ABCB5. Todos los melanomas tuvieron una expresión variable de ABCB5 detectado por hibridación in situ, lo cual sugiere que el gen puede ser regulado diferencialmente en melanomas individuales.
ConclusionesEl gen ABCB5 podría estar relacionado con las propiedades de resistencia a la quimioterapia y la agresividad del melanoma. La elevada expresión encontrada en las muestras de melanoma acral podría ayudar a la mejor comprensión de la patogenia de esta forma frecuente de melanoma en la población mexicana, que se asocia generalmente con un peor pronóstico.
The incidence of melanoma is increasing at a rate faster than that of any other cancer, and its mortality rate continues to increase in some parts of the world. This highlights the need for improved methods of diagnosis and treatment as well as a better understanding of some types of melanoma that have been scarcely studied, such as acral melanoma (AM). This is a frequently misdiagnosed form of melanoma that develops on palmar, plantar and subungueal skin.1
Melanoma is characterized by a high risk of metastasis, which once it occurs implies a low survival expectancy (7–12 months).2 In the progression of metastasis, the molecular events that form a complex process and involve multiple genes take place sequentially.3
Previous studies of cutaneous melanoma have found alterations in several genes such as N-RAS, H-RAS, FGF and PTEN.4 It has also been observed that metastatic melanoma displays resistance to antineoplastic drugs.5 The causes of chemotherapy failure range from the physical inability of the drugs to reach their cellular target to diverse cellular and molecular mechanisms of resistance.6 This phenotype may be due to the P-glycoprotein and some membrane transporters of drugs associated with resistance to a broad spectrum of lipophilic drugs.7,8 A very important finding in drug resistance research is the knowledge of the phenomenon known as multidrug resistance (MDR), whereby exposure to one drug induces and/or makes evident cross-resistance to a variety of agents to which the cell has not been exposed.6,9,10 One of the main mechanisms that may mediate MDR is the expression of proteins that act as a transmembrane exporter of drugs.6,9–11 Although the overexpression of the P-gp is important in MDR, there are two other related factors: (a) genomic amplification and (b) polymorphic sequence variation. The sequence variations of this membrane pump have different capacities to bind with and expel drugs from the cytoplasm. In human cancer cells, it has been reported that the P-gp (ABCB1) gene shows genomic amplification and polymorphic sequence variations.12 Although the molecular chemoresistance mechanisms are not yet completely understood, several molecules have been implicated such as the families of MDR pumps, topoisomerase II and some antiapoptotic molecules.13
Our work describes a molecular screen for identifying differentially expressed genes in acral melanoma, which is the most common melanoma in the Mexican population.2 We used differential display reverse transcription-polymerase chain reaction (DDRT-PCR), which is a valuable technique for identifying mRNAs that are expressed in different conditions (cancerous versus healthy tissue), stimulus or cells of similar background and are compared at the same time. Differential display (DD) was created by Liang and Pardee14 to allow a rapid, accurate and sensitive detection of altered gene expression. This technique works by amplification of the 3′ terminal portions of mRNAs and resolution of those fragments on a DNA sequencing gel. Using anchored primers designed to bind the 5′ boundary of the poly-A tails for the reverse transcription, followed by PCR amplification with additional upstream primers of arbitrary sequences, mRNA subpopulations are visualized by denaturing polyacrylamide electrophoresis. In addition, it constitutes an important tool for scientists interested in the identification of novel genes coding for key proteins that play a major role in diseases such as cancer, cerebral ischaemia, etc. One of the advantages of DD is the capacity to use multiple lanes to study several RNA populations simultaneously from small amounts of starting material.
We compared the gene expression patterns between primary melanomas and normal skin,15 finding that the ABCB5 gene was expressed differentially in one sample of melanoma tissue. When using RT-PCR, the bands of PCR amplification of the ABCB5 gene were detected in nine out of ten fresh acral melanoma biopsies.
ABCB5, a human ATP-binding cassette transporter, has been recently implicated in the regulation of cell fusion, and the resultant growth and differentiation of progenitor cells.16 As previously suggested, the new ABCB5 gene may be related to the properties of chemoresistance and malignancy in melanoma.11,16,17 However, ABCB5 gene expression detected by in situ hybridization (ISH) has not yet been reported in any of the multiple studies of primary melanoma tissues.
Materials and methodsThis study was approved by the Institutional Research and Ethics Committee and informed written consent was obtained from all subjects.
Melanoma tissuesFor the differential display (DD) study, ten primary AM samples were obtained from Mexican patients who underwent surgery at Hospital General de México, in México City. All patients had regional lymph node metastasis and a melanoma with a median Breslow thickness of more than 1cm (Table 1). All tumors were classified as AM by both a clinical oncologist and an expert dermatopathologist. A sample of normal skin was obtained from every patient as a control. For the DD analysis, part of the tissue was frozen at −70°C and the rest was used for histopathological studies.
Clinical data of the patients with different types of melanoma and the expression of ABCB5 and tyrosinase
| Case | Age | Sex | Breslow | Clark | Type | ABCB5a | TYRa |
| 1 | 30 | F | – | – | SSM | 0.01 | 3.8 |
| 2 | 66 | M | 0.92mm | III | SSM | 0.14 | 4.54 |
| 3 | 83 | F | 0.7mm | III | SSM | 1.0 | 4.42 |
| 4 | – | M | 0.56mm | III | SSM | 0.04 | 0.71 |
| 5 | 81 | M | 0.1mm | I | SSM | 1.58 | 2.04 |
| 6 | 23 | F | 0.9mm | III | SSM | 0.4 | 2.9 |
| 7 | – | M | 0.45mm | IV | SSM | 1.0 | 3.2 |
| 8 | 58 | M | 0.45mm | II–III | SSM | 0.3 | 0.01 |
| 9 | 72 | F | – | – | AM | 1.4 | 3.9 |
| 10 | – | F | 5mm | V | AM | 1.14 | 6.13 |
| 11 | 72 | M | >1cm | VI | AM | 6.02 | 2.0 |
| 12 | 59 | M | >1cm | VI | AM | 5.98 | 4.2 |
| 13 | 67 | M | >1cm | VI | AM | 1.31 | 1.574 |
| 14 | 48 | M | >1cm | VI | AM | 1.0 | 2.1 |
| 15 | 73 | M | >1cm | VI | AM | 5.3 | 1.61 |
| 16 | 66 | M | >1cm | VI | AM | 8.0 | 5.4 |
| 17 | 69 | F | >1cm | VI | AM | 9.04 | 4.78 |
| 18 | 60 | M | >1cm | VI | AM | 1.35 | 2.04 |
| 19 | 72 | F | >1cm | VI | AM | 3.63 | 0.48 |
| 20 | 55 | F | >1cm | VI | AM | 6.2 | 7.5 |
| 21 | 57 | F | >1cm | VI | AM | 2.7 | 0.9 |
| 22 | 59 | F | >1cm | VI | AM | 2.0 | 5.9 |
AM: acral melanoma; F: female; M: male; SSM: superficial spreading melanoma.
The fluorescence emitted by ISH using the ABCB5 probe as well as the tyrosinase probe was measured in pixels per area, based on the evaluation of three melanocyte cells per viewing field of healthy skin with the Image J® program. The results of this evaluation were the basis for comparing the data obtained in all the analyses of acral melanoma and superficial spreading melanoma.
Twenty two melanomas were included in this study: seventeen paraffin samples (eight from superficial spreading melanoma, nine from AM) and five fresh melanoma tissues were used for the DD technique.
Differential display RT-PCRTotal RNA was isolated using the RNA aqueous PCR kit (Ambion, Austin, TX, USA). Ten milligrams of tumoral tissue was pulverized with liquid nitrogen and later processed as indicated by the manufacturer's instructions. Total RNA was treated with DNase I, and mRNA differential display was performed as described.14 First strand cDNA was synthesized using 25pM of HT11A primer (Genhunter Corp., Nashville, USA) and 1μl (40U) of Superscript II enzyme (Invitrogen, New York, USA).
Samples of each reverse transcription were amplified by PCR using the same oligo (dT) primer used for first-strand cDNA synthesis and different arbitrary oligonucleotide 10 mer primers (5pM) and 1U of Taq polymerase (Invitrogen, New York, USA).
Twenty four primer combinations from Gen Hunter RNA image kit were used as randomly selected 5′ primers. PCR conditions were 94°C/2min (94°C/30s, 40°C/2min, 72°C/30s) for 42 cycles and a final extension of 7min at 72°C. All cDNAs from melanoma tissues were done in a duplicate manner to ensure reproducibility. The samples were run on ultra-thin gels and stained with the silver nitrate method as the manufacturer recommends (Cleangel DNA analysis kit, Amersham, NJ, USA).
Reamplification of candidate bandsThe differentially expressed bands were identified by ocular inspection and the bands of interest were excised from the gel (using an independent scalpel blade for each sample), boiled in water (10μl) and reamplified using 1U of Taq polymerase (Invitrogen, New York, USA), 100μM of dNTPs (dATP, dCTP, dTTP and dGTP) and PCR buffer to a total volume of 10μl. The reamplification conditions of PCR cycles were the same as those of the conventional PCR, and the amplified tubes were diluted 1:20.
Cloning and sequencingReamplified cDNA fragments were cloned into the pGEM vector from Promega (Madison, WI, USA) according to the manufacturer's instructions. The cDNAs were sequenced using the Sequencer ABI Prism 377 DNA with the Dig Dye kit (Applied Biosystems, California, USA).
The sequences of the isolated cDNA clones were determined by computer search and compared with the Gene bank data bases.
RT-PCRABCB5 expression was evaluated by RT-PCR using mRNA from ten fresh AM tissues. Primers for exon 4 and 5 of the ABCB5 gene were designed as follows, forward: 5′-CAGAGCTTTAAATGTGCGGC-3′ (ABCB5 cDNA positions 488-507); reverse: 5′-TACCACGATTGTAGTCCGAC-3′ (ABCB5 cDNA positions 850-869).
Amplification conditions were 94°C/5min, followed by 38 cycles (94°C/30s, 57°C/30s and 72°C/30s). There was a final extension of 7min at 72°C. Beta actin primers were used as house keeping gene control. Amplification conditions were the same as those reported by Kurebayashi et al.18
In situ hybridizationTissue slices were mounted onto slides with poly-l-lysine (1:10) (Sigma-Aldrich Co., USA).
Xylol treatment was used for deparaffination, and rehydration was carried out with alcohol–water (100%, 95%, 70% and 25%). Samples were permeabilized using proteinase K (3μg/ml) and were then incubated in 1% paraformaldehyde for 10min. Prehybridization was done with 50% Hybrizol (Chemicon International, California, USA) and 50% formamide at 40°C for 1h.
Hybridization was done using 500ng of each probe, at 80°C/5min and 40°C O/N. Two washes were done with SSC 2× at room temperature for an hour and two more with SSC 0.2× at 40°C for an hour. Vectashield® (Vector Laboratories, Burlingame, CA, USA) was used to mount tissues on slides. Probes used for ISH were reverse for ABCB5: (5′-TACCACGATTGTAGTCCGAC-3′) labeled by Cy5; reverse for TYR (5′CTACAGACAATCTGCCAAGAGGAG-3′) labeled by fluorescein (cDNA position 889-912) and control forward for ABCB5 (5′-CAGAGCTTTAAATGTGCGGC-3′) labeled by fluorescein.
Laser scanning confocal assaysLabeled specimens were scanned with a LSM-5 Pascal confocal laser-scanning microscope (Carl Zeiss, Jena Germany) linked to a Zeiss inverted microscope Axiovert 200M equipped with a Zeiss 100× plan-neofluar oil immersion lens with a numerical aperture of 1.3 (Carl Zeiss, Jena Germany). The 488nm line from an argon laser was used for the fluorescein signal and the 633nm line from a helium–neon laser was used for the Cy5 signal. Fluorescence signals from fluorescein were observed using a dichroic beam splitter (HFT 488/543/633; Carl Zeiss) and an emission filter (BP505-530; Carl Zeiss). Optical fluorescence signals of Cy5 were observed using a dichroic beam splitter (HFT 488/543/633; Carl Zeiss, Jena Germany) and an emission filter (LP650; Carl Zeiss, Jena Germany). A computer equipped with KS-300 software version 3.0 (Carl Zeiss, Jena Germany) was used for operating the system. Detector gain and pinhole aperture were automatically adjusted. The image resolution was 1024×1024 pixels (8 bits, 256 color levels). Experiments were performed in triplicate and repeated independently three times.
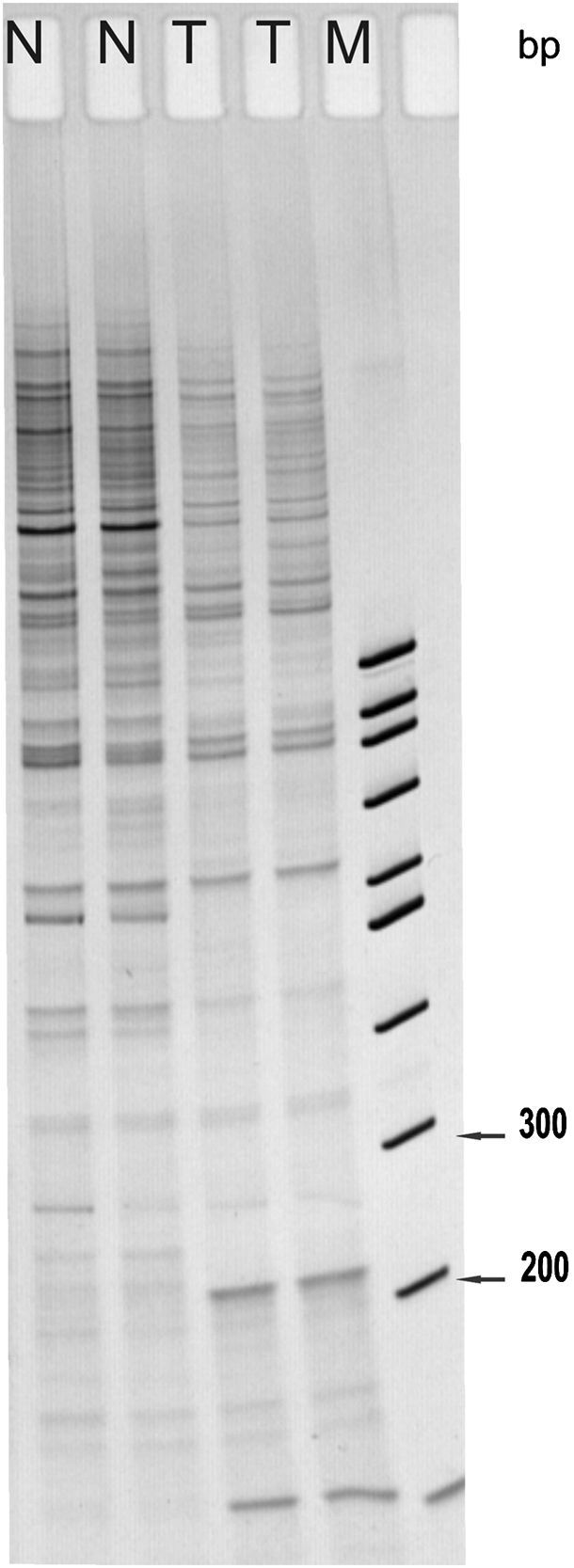
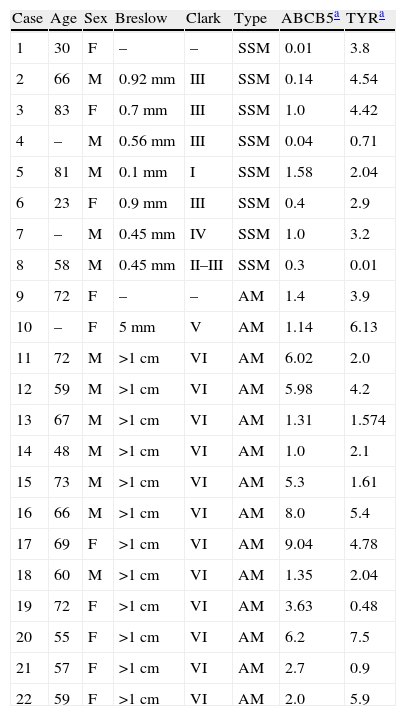
ResultsA 258bp band was expressed differentially by DD-PCRThe present study describes differential patterns of gene expression in normal skin versus AM detected by mRNA DD with different 5′ primers in combination with sets of 3′ primer pools. A representative example of DD results is shown in Figure 1. Although we also found other differentially expressed genes, they are not the focus of this study. A 258bp de novo band detected by decameric primer combination (H-T11VA)14,15 was expressed in an AM tumor but not in normal skin.
DD-PCR applied to malignant melanoma mRNA and normal skin, using the primers H-T11G, A, C (Gen Hunter) and 20 arbitrary 10mer primers. The PCR products were separated on ultra-thin gels and stained with Silver Staining Kit. Lanes N are normal skin and lanes T are acral melanoma (patient 14 of Table 1). A 258bp de novo expression band (arrow) and a 100bp ladder molecular marker in lane M are shown.
The specific band was reamplified, cloned and sequenced as described in the Materials and Methods section.
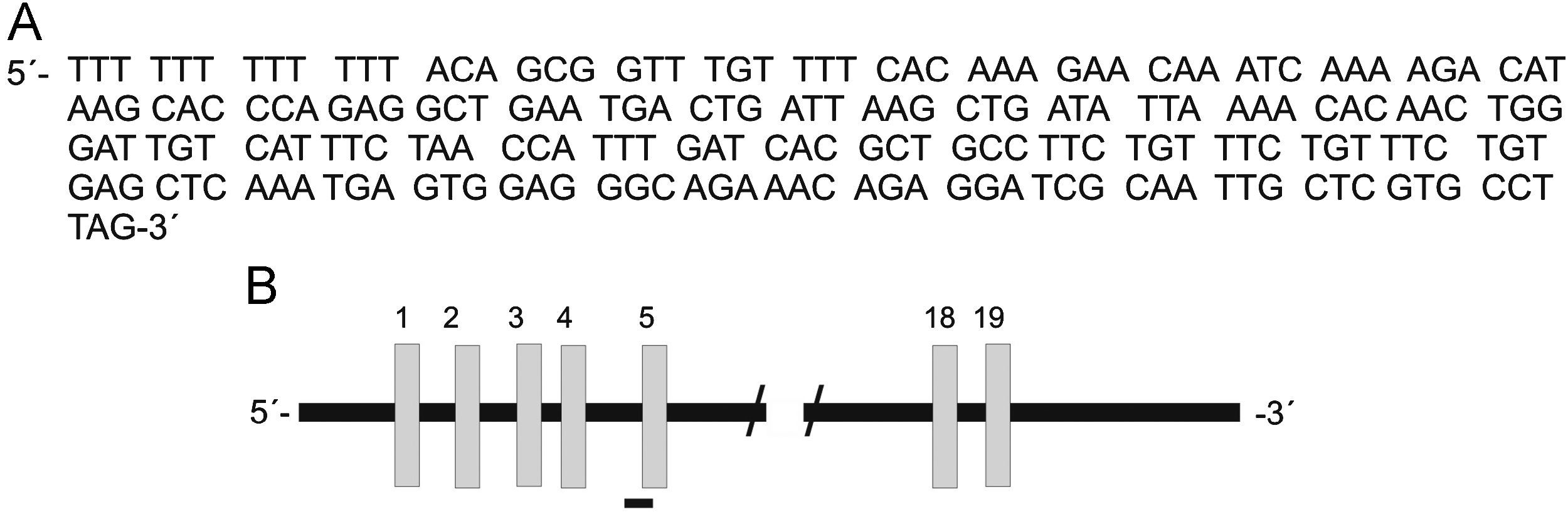
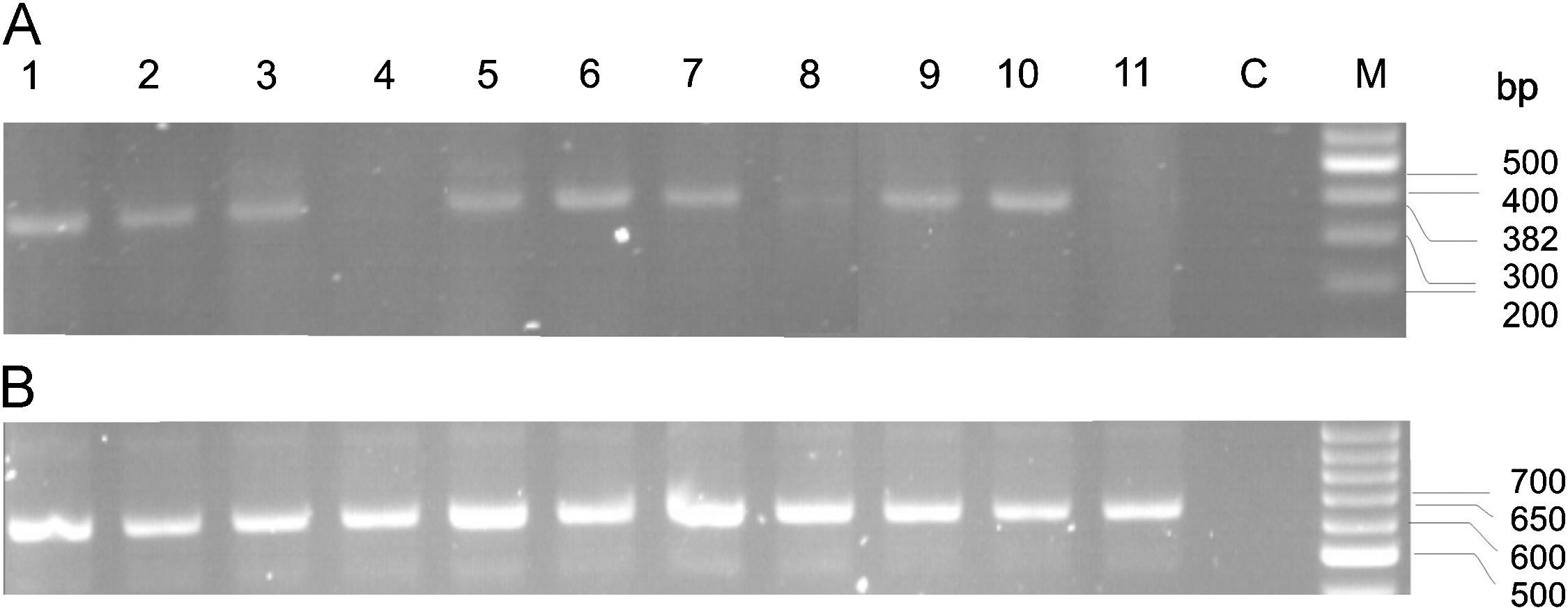
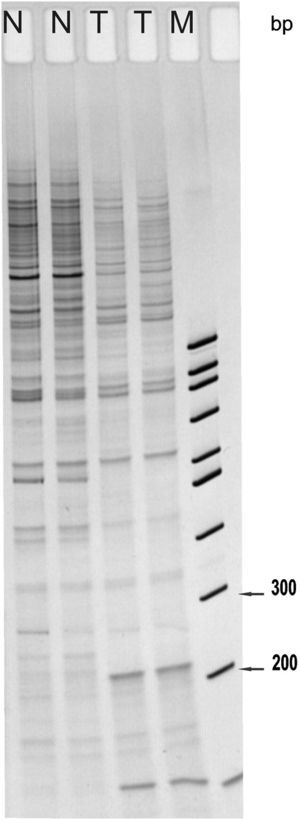
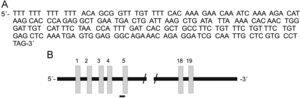
The sequence was analyzed by the BLAST NCBI program and showed 100% homology to the ABCB5 gene, an ATP-binding cassette transporter encoded on chromosome 7p21-15.3 (Figure 2). This gene has been implicated in the regulation of cell fusion and the resultant growth and differentiation of progenitor cells.16 The 258bp fragment found was located between intron 4 and exon 5 of the gene. Primers located in this sequence of the ABCB5 gene were designed for RT-PCR detection. The ABCB5 mRNA gene was detected (9/10; all rows but 4) in 90% of the tumor samples (Figure 3A). The beta actin mRNA control was amplified in all tumors as well as the control tissue (Figure 3B).
The 258bp differential band was cloned and sequenced by using an automatic capillary sequencer. The sequence was found to be 100% homologous to the ABCB5 human gene. The fragment is located between intron 4 and exon 5 of the genomic sequence. Bottom diagram shows the genomic map of ABCB5 and the DD fragment location.
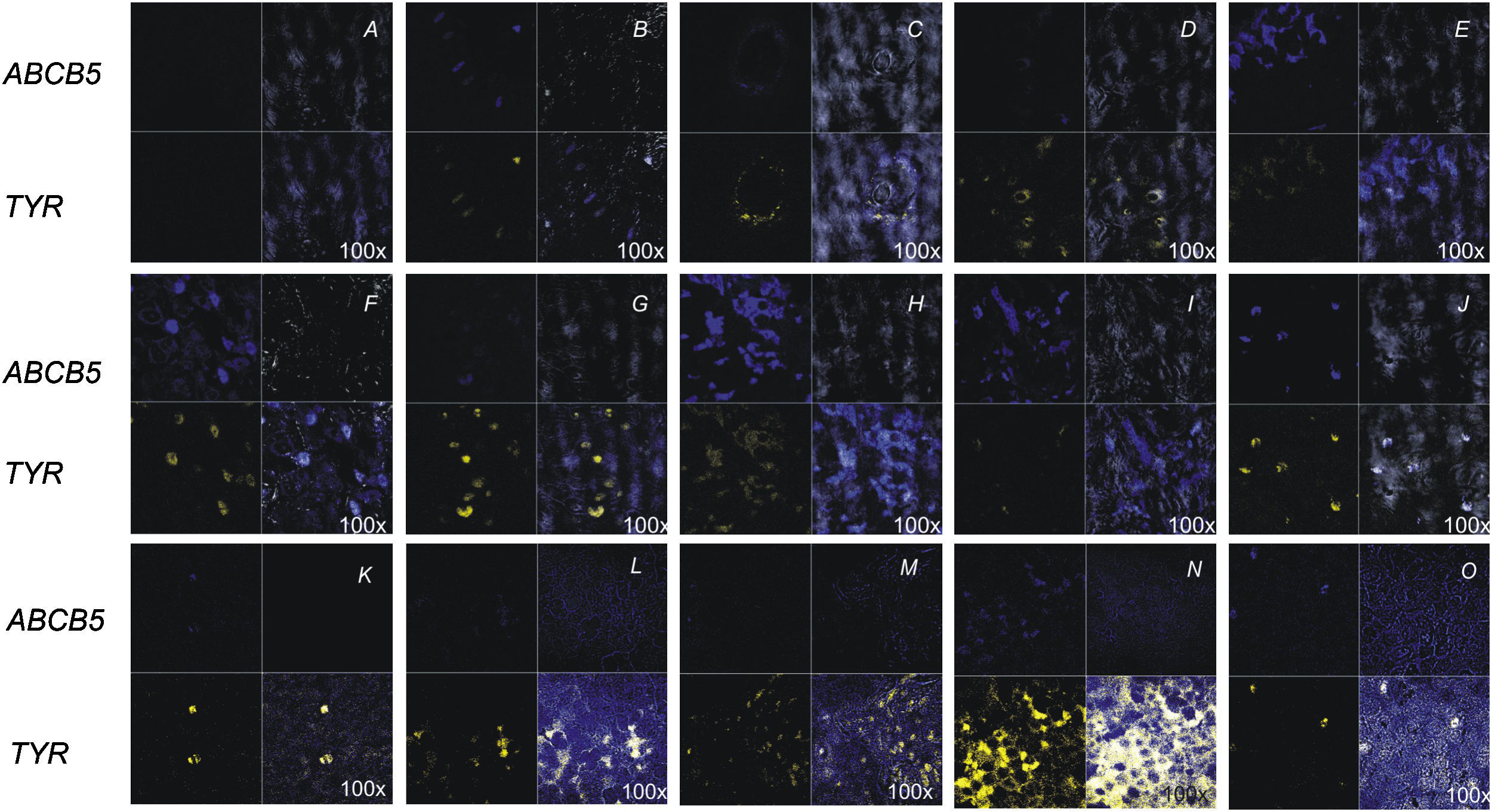
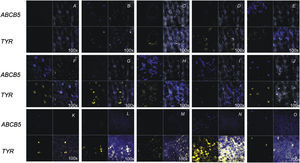
Clinical characteristics of the patients for the 22 samples are summarized in Table 1. The ABCB5 gene was detected using an ISH study and specific fluorescent probes (Cy5, 5′-labeled). ABCB5 mRNA was detected in 100% of the melanoma tissues, although with different intensities depending on the sample. These samples and the normal skin were also hybridized with a tyrosinase mRNA control and analyzed with confocal microscopy software (Figure 4). Although tyrosinase has been proposed as a marker of melanomas, its expression in this study was found to be quite variable, in some cases lower than in healthy skin.
In situ hybridization of melanoma tissues for ABCB5: The ABCB5 probe was labeled with Cy5 (blue) and tyrosinase probe was labeled with fluorescein (yellow), and both were observed by confocal microscopy. For A–J, up left Varell®; up right ABCB5 probe; down left merge; down right tyrosinase probe. A) Negative control (probe ABCB5 sense). B) Normal skin. C) Transversal section of skin follicle. D) SSM (patient 2). E) Superficial spreading melanoma (SSM) (patient 5). F) Acral melanoma (AM; patient 11). G) AM (patient 13). H) AM (patient 14). I) AM (patient 18). J) AM (patient 15). K) AM (patient16). L) AM (patient 17). M) AM (patient 20). N) AM (patient 22). O) AM (case 19) (100× magnification).
It is interesting that in the indigenous and Mestizo people of México, AM is the most common (75%) form of melanoma and it is usually detected as an already thick tumor in glabrous skin areas.2,19 These anatomic regions have a very low exposure to sunlight and are protected from UV radiation by a thick corneum stratum, which raises the question as to the possible mechanisms, such as genetic background,20,21 that could be involved in the development of AM.
The molecular mechanisms involved in AM are largely unknown.19 Bastian et al.4,22,23 showed in various studies that AM is different in type from superficial spreading melanoma, which is the most common type in Caucasians and is characterized by genomic amplifications occurring early in tumorigenesis.4,22,23 Davies et al.24 identified 66% of activating mutations of BRAF (V600E) in melanoma cell lines. B-RAF is a member of the RAF family, which encodes serine/threonine kinases that act in the MAKP pathway to transduce regulatory signals from Ras to MEK.24,25
In the current study different levels of gene expression were found in the samples of normal skin versus those of AM and SSM. The melanomas studied by DD were from patients that had regional lymph node metastases, implying low survival expectancy (7–12 months). These differentially expressed genes might be related to melanoma progression, as Schatton proposed in his recent work.11 We found the ABCB5 gene highly expressed in 90% of the samples (9/10) of fresh AM tissues by using RT-PCR. This gene has been recently described by Frank et al16 and has been implicated in the regulation of progenitor cell fusion (located in chromosome 7p21-15). Their study used an enriched culture of human epidermal melanocytes isolated from the foreskins of healthy donors and from melanoma cell lines.
Seven human MDR subfamily genes (ATP binding cassette) from A to G are known, in which the expression control occurs at the level of the gene copy number, transcription, translation and post-translation.9,26 The MDR genes are conserved across the species, which suggests a relevant role of their products in the survival of the organism.9
The ABCB subfamily includes 11 members that have different expression patterns.9 Whereas the functions of several transporters of the ABCB family are unknown, there is some information, albeit very limited, about the specific case of ABCB5 in relation to its pattern of expression or its associated function.6
The main characteristic of the ABCB5 is the predicted extracellular amino acid loop that is probably related to the fusogenic properties of the melanocyte precursor cell.16 Two transcript isoforms have been described (alpha and beta), which are splicing products.27 This study showed that when siRNA is directed to ABCBα, it is possible to reduce the ABCB5 mRNA level. This induces drug sensitivity to camptothecin, 10-OH, 5-FU and methotrexate, among others, thus reverting the MDR phenotype.15,30
Frank et al.28 studied melanoma cell lines and showed that the ABCB5 is the main protein that participates in doxorubicin chemoresistance. If the pump is blocked by using specific monoclonal antibodies, it is possible to revert the MDR phenotype. P-gp also plays a role in MDR in sarcomas and leukemia.26 However in other malignant tumors, such as melanoma, chemoresistance and radioresistance processes are the result of complex mechanisms that involve different molecular strategies such as grade progression, clinicopathological behavior and host characteristics.27 In breast cancer studies, there is evidence that P-gp plays a role in chemoresistance to cytotoxic drugs.9,29 In one of these studies, it was emphasized that P-gp expression was found more often in metastatic rather than in primary tumors, and in the latter was associated with the presence of three or more positive auxiliary nodes.6
Weinstein et al.30 suggested that MDR expression in colorectal cancer should be regarded as a molecular indicator of tumor aggressiveness. They reported that half of the patients with primary tumors did not express P-gp and more than half of metastatic cells show P-gp expression. They suggest that P-gp may influence the behavior of these cells.
Chen et al.31 suggested an interesting hypothesis regarding the MDR phenotype of melanoma. They demonstrated that this characteristic involves the subcellular removal of intracellular cytotoxic drugs by the melanosomes as well as increased melanosome-mediated drug extrudability.
Melanoma is a heterogeneous malignant disease characterized by four main histopathological types: acral, nodular, superficial spreading and lentigo maligna melanama (AM, NM, SSM and LMM, respectively). Among the molecular differences between them, there are genomic amplifications and differentially expressed genes in AM tumors, point mutations (BRAF, V600E) in SSM and deletions that involve the 1p36 chromosome in 63% of NM.32,33
Human cancer distribution is influenced by genetic background, local habits and environment. In different ethnic groups, the clinical behavior and natural history of some diseases show variations that are ultimately an important factor in their pathological progression. While the Mexican population includes different ethnic groups, the skin phototypes III and IV are the most prevalent. Compared to Caucasians, SSM has a low prevalence in the Mexican population. Melanin in melanosomes forms a shield to the UVB radiation and protects the genomic integrity of keratinocytes and melanocytes.1
The first two molecular studies of ABCB5 in melanoma were done by Frank et al.,16,28 where they used HEM, cell lines and some melanoma samples. Their results, using monoclonal antibodies, identified ABCB5 as a novel drug transporter. Our DD-PCR study was performed with melanoma primary tumors of Mexican patients. In the ISH assays we show that there are qualitative and quantitative differences in ABCB5 gene expression between samples of melanoma that were different in their stages and histological patterns (Figure 4). Results show that even though the ABCB5 gene is expressed in most of the melanomas (regardless of the type), in AM the expression of this gene is detected in a higher proportion and more intensely than in SSM (data not shown). We found a direct relationship between the expression of mRNA for ABCB5 and the degree of pathogenicity of acral melanoma. However, in intermediate stages of superficial spreading melanoma, the expression was lower. Also worth noting is that the expression of ABCB5 in the cases of AM 15–17 was found to be more than 5 times that observed in healthy skin.
It has been showed that the polymorphic variations of this gene and its degree of expression can alter the response of a patient to chemotherapy. The modification of this response in different patients would be an interesting field for future studies of ABCB5 polymorphism.34,35 It should also be very interesting to study the expression of this gene in two groups of melanoma patients: those that respond to chemotherapy and those that do not respond and progress to metastases.
The discovery of the mechanism that is responsible for eliminating antineoplastic drugs from cells has opened up a new area of research, which is beginning to characterize this novel mechanism in relation to the degree of cancerous invasion and the clinical behavior of the tumor.11,36
FundingThis research was supported by the following grants: CONACYT (no. 38450-M), Secretaría de Investigación y Posgrado, IPN. (SIP no. 20050772) and SIP (no. 20060731) for PhD to N.E. Herrera-González.
Conflict of interestAuthors have no conflict of interest to declare.