X-linked hypohidrotic ectodermal dysplasia (XLHED) is characterized by abnormal development of the hair, teeth, and sweat glands. It is caused by mutations in the EDA gene, which maps to the X chromosome and encodes a protein called ectodysplasin-A, a member of the tumor necrosis factor-related ligand family. Affected males typically exhibit all the typical features of HED, but heterozygous carriers may show mild to moderate clinical manifestations. We describe the case of a Spanish family in which a novel heterozygous c.733_734insGA mutation at the EDA gene was identified. It was located in exon 5 and consisted of a frame-shift mutation at codon 245, which gave rise to an abnormal protein with a premature stop codon after 35 residues. Genetic analyses in families with XLHED are useful for checking carrier status, but they also provide information for genetic counseling and prenatal diagnosis.
La displasia ectodérmica hipohidrótica ligada al cromosoma X (XLHED) se caracteriza por un desarrollo anormal del pelo, los dientes y las glándulas sudoríparas. Está producida por mutaciones en el gen EDA, que se localiza en el cromosoma X y codifica para la proteína Ecdodisplasina A, miembro de la familia de ligandos relacionados con el factor de necrosis tumoral. Los varones afectados normalmente exhiben todas las características de la enfermedad, pero los portadores heterocigotos pueden mostrar manifestaciones de leves a moderadas. Aquí se describe una familia española en la que hemos identificado una mutación c.733_734insGA, previamente no descrita, en el gen EDA. Se localizaba en el exón 5 y producía un cambio en la fase de lectura en el codón 245 de la proteína, lo que daba lugar a un codón de parada prematuro tras 35 residuos. El análisis genético en familias con XLHED es fundamental para la identificación de las portadoras, para el diagnóstico prenatal y en general para proporcionar un asesoramiento genético correcto.
X-linked hypohidrotic ectodermal dysplasia (XLHED), OMIM 305100, is a rare genodermatosis characterized by an abnormal development of the hair, teeth, and sweat glands. XLHED is the commonest form of hypohidrotic ectodermal dysplasia, accounting for 95% of cases. It is found worldwide, with an estimated incidence of 1 in 100,000 births. It is caused by mutations in the EDA gene (also called ED1), which maps to the X chromosome (locus Xq12-q13) and it encodes a protein called ectodysplasin A, a member of the tumor necrosis factor-related ligand family that is probably involved in the early epithelial–mesenchymal interaction that regulates formation of the ectodermal appendages.1
Affected males normally exhibit all the typical features of HED, but heterozygous carriers may show mild to moderate clinical manifestations.2 We describe the findings in a Spanish family with XLHED with a novel pathogenic mutation in the EDA gene.
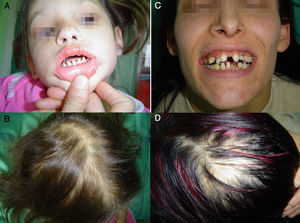
Patients and methodsThe proband (III-5) was a 4-year old girl who had pointed teeth, sparse thin hair, and a congenital scarring lesion on the bridge of the nose, which was thought to be a Blaschko line at that site (fig. 1A, 1B). The tail of the eyebrows was absent bilaterally, her skin was dry, and sweating was reduced. She also presented a prominent lower lip and hyperpigmentation and dryness around the eyes. Her nails were normal. The results of routine laboratory analyses were within the normal range. Her family stated that she had suffered from febrile convulsions soon after birth, although medical advice had not been sought. Her mother (II-3), who was 35 years of age, presented similar clinical features to the proband (III-5) (figs. 1C, 1D), although her skin was not so dry and her sweating could be described as being about normal. Her hyperpigmented lesions were more prominent and she also exhibited some eczematous plaques. In addition, she had never developed her adult dentition. The grandmother (I-2) and the grandfather (I-1) of the proband did not display phenotypic features, nor did the proband's father (II-4). Genetic analysis of the grandfather could not be performed as he had died. The pedigree is shown in Figure 2A.
A and B. Clinical features of the proband. C and D. Clinical features of the proband's mother. The proband has pointed teeth, absence of the tails of the eyebrows, a congenital scaring lesion on her nose (A), and sparse thin hair (B). Her mother exhibits similar clinical signs (C and D). She never developed her adult dentition.
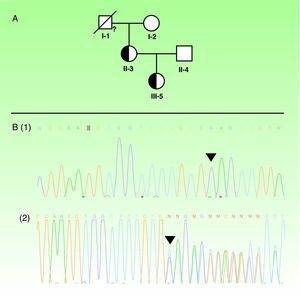
A. Pedigree of the family analyzed. The proband (III-5) is marked with an arrow. Half-filled symbols represent affected heterozygous carriers. A quotation mark indicates the members that were not analyzed genetically. B. The EDA mutation is shown: (1) the normal sequence; (2) the picture of the heterozygous; c. 734_735insGA mutation in the EDA gene. The black triangle indicates the site of mutation.
After giving their informed consent, genomic DNA was obtained from peripheral blood from the proband and from her family members. PCR amplification of the EDA gene obtained from the DNA samples was performed. The PCR conditions and primers used to amplify the EDA gene have been described previously.3 The reaction was conducted in a 2720 Thermal Cycler (Applied Biosystems®). Analyses were performed in duplicate to ensure reliability of the results.
Results and discussionDirect sequencing revealed a novel heterozygous c.734_735insGA mutation in the EDA gene in the proband and in her mother (fig. 2B). The grandmother and the father of the proband did not carry the mutation. One hundred alleles from healthy people were tested and failed to reveal that mutation, which was located in exon 5 and consisted of a frame-shift mutation at codon 245. This gave rise to an abnormal protein with a premature stop codon after 35 residues (Asn280Ter), generating a protein lacking the TNF homology domain. The different mutations identified in the EDA gene are located particularly in 3 functional clusters: the furin protease recognition site, whose codon 156 is the one most frequently affected; the collagen-like domain; and the TNF homology domain itself, as seen in this family.4 One hundred sixty-one pathogenic mutations have been recognized in XLHED (Human Gene Mutation Database, 25th March 2011). To our knowledge, the mutation we describe has never been reported.
The impact of this mutation can be appreciated by means of a brief review of protein structure and function. In general, the EDA gene gives rise to several different transcripts through alternative splicing. The longest transcripts, EDA-A1 and EDA-A2, are the most relevant ones from the biological point of view.5 These transcripts are produced as trimeric type II transmembrane proteins that are released from the cell surface by furin-mediated proteolytic cleavage. TNF domains are of great relevance for this protein, and their absence, as seen in our patients, leads to protein dysfunction due to an inability to bind the receptor. For further information about the role of EDA in the development of skin appendages and in NF-kB signalling pathway activation, readers are referred to reference.6
The proband exhibited a more severe phenotype than her mother, explained by the X inactivation process. Because of this mechanism, some XLHED female carriers may exhibit a very mild phenotype and the probability of detection by clinical examination alone has been estimated to be of 60 to 70%.2 The XLHED carrier status can only be unequivocally determined by molecular analysis. A female carrier (the proband and her mother) has a 50% probability of having a male child with clinically severe XLHED. This observation is of particular value in the context of genetic counseling.7
This report expands our knowledge of mutations in the EDA gene. Mutation analysis in families with XLHED enables carrier status to be determined and genetic counseling and prenatal diagnosis to be performed.
Conflict of interestThe authors declare that they have no conflict of interest.