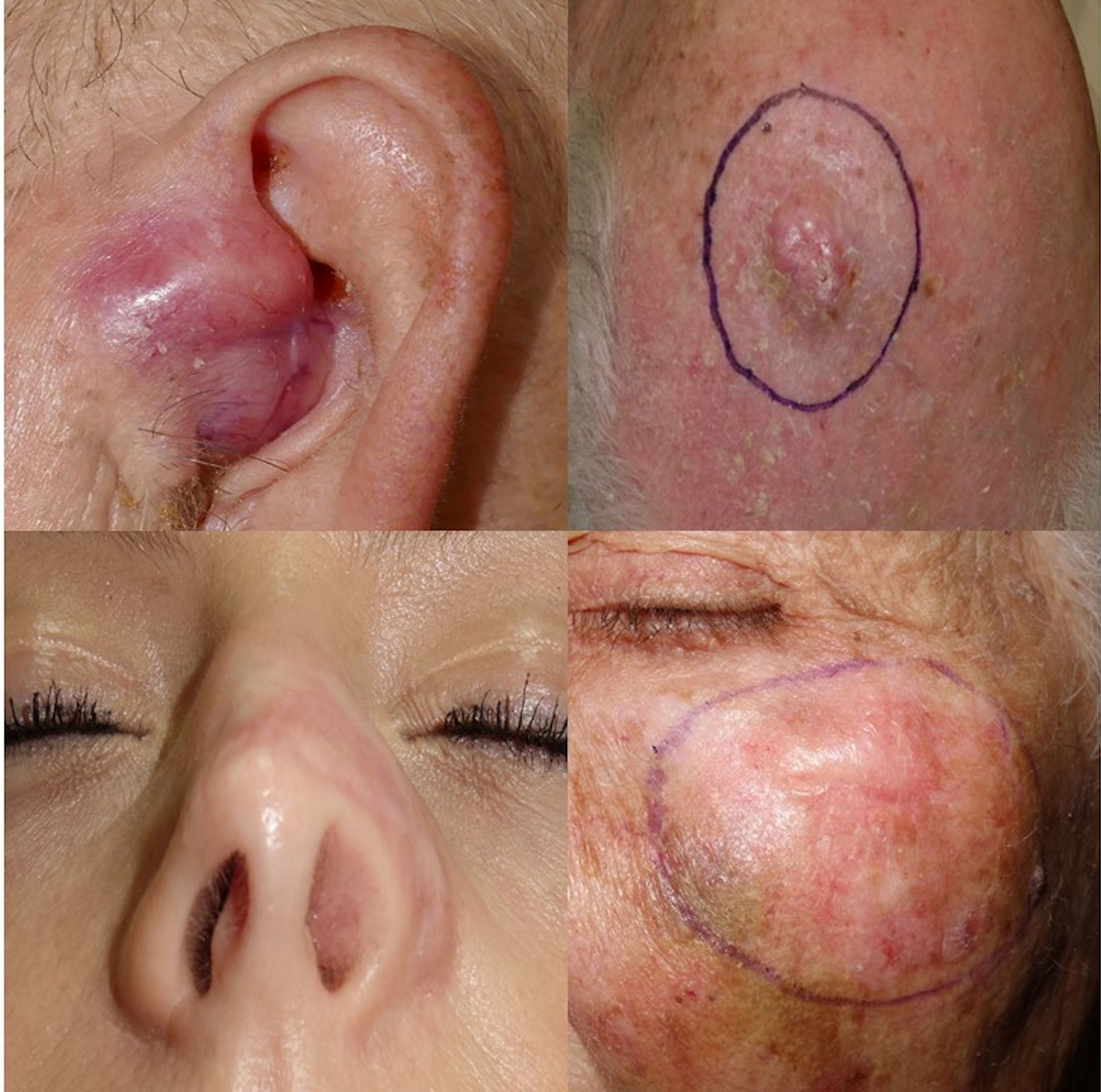
Desmoplastic melanoma (DM) is a rare subtype of melanoma that affects patients older than 70 years with intense sun damage. DM shows a good response to immunotherapy and is characterized by a high mutation rate.1–3
We studied a total of 12 cases of DM and analyzed the clinical and histopathological variables. Mutational studies were performed through massive sequencing of all tumours. DNA and RNA were amplified with the Oncomine Precision Panel – GX5 – Solid Tumour w2.6.0 DNA and Fusion Panel (Thermo Fisher Scientific), generating a library and including amplicons for the study of mutations and INDELs in the hotspot regions of 45 genes, copy number variations in 14 genes, and fusions in 18 genes. Sequencing was analyzed on the Genexus System platform.
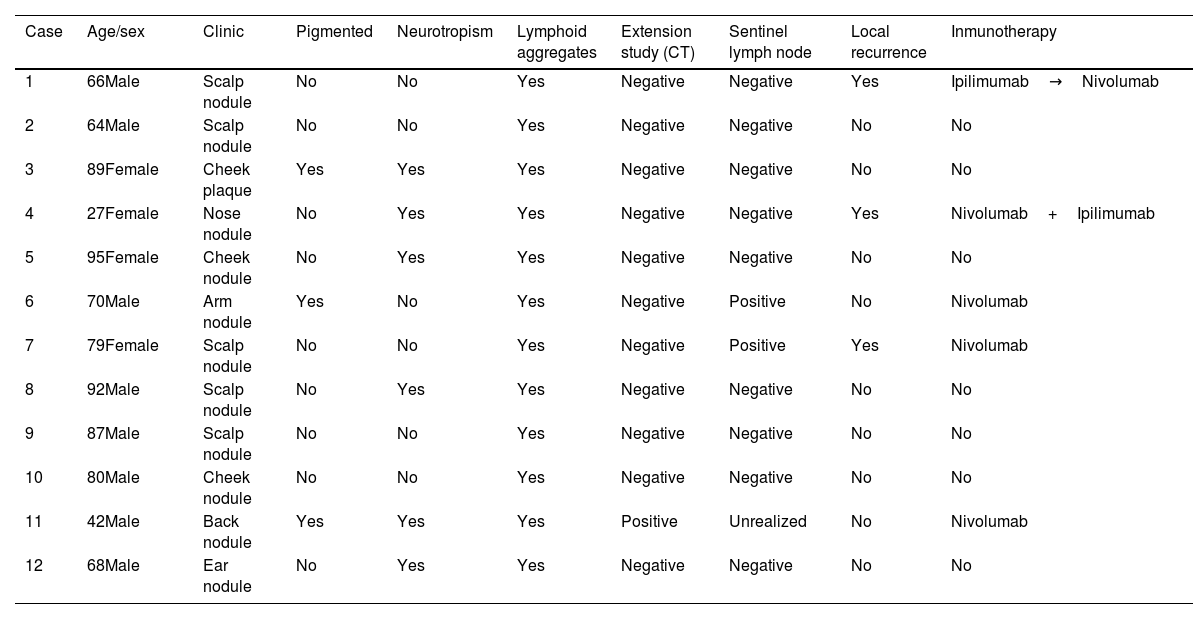
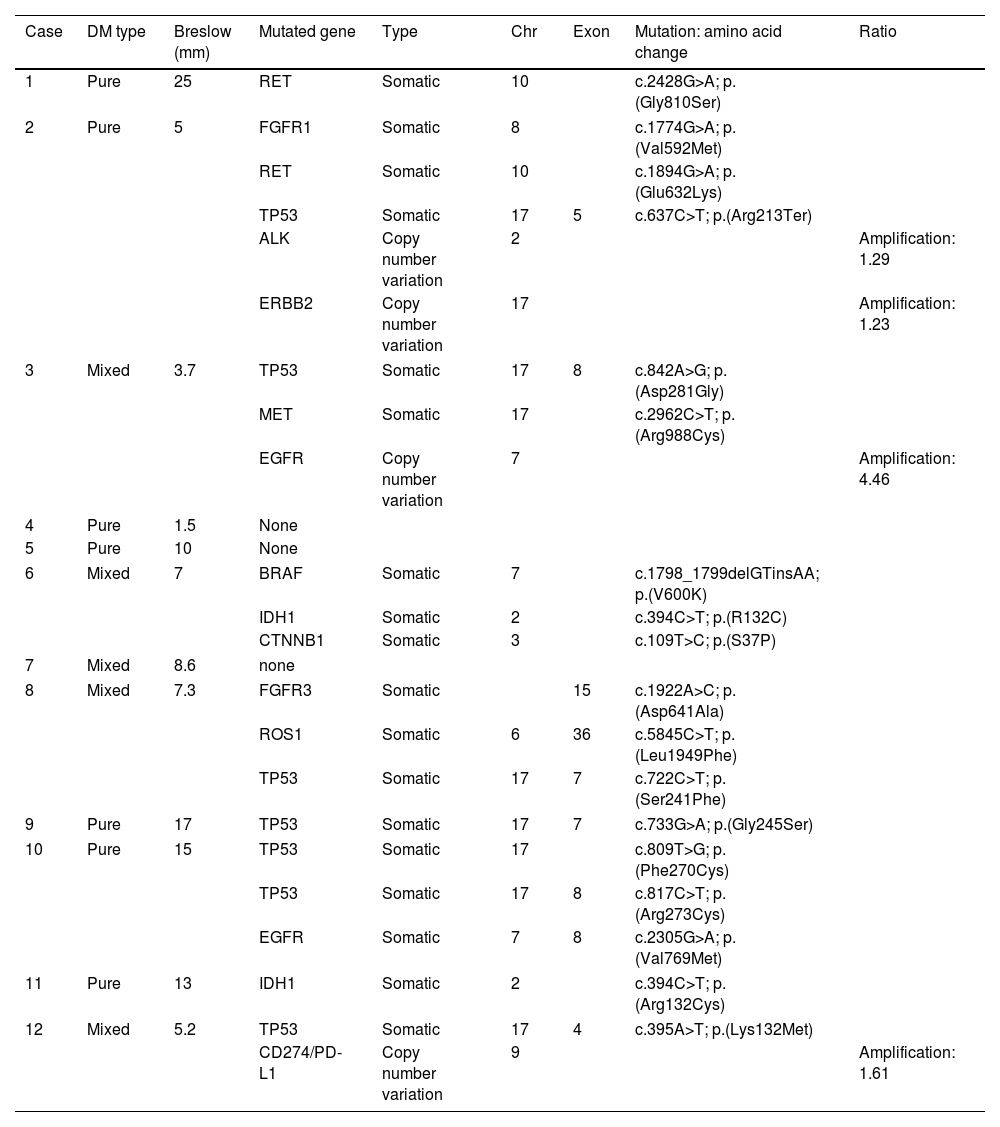
A total of 12 patients (8♂:4♀) with a median age 70 years and diagnosed with DM were studied. The most common presentation was nodules on the scalp (5 cases, 41.6%), with other common presentations being nodules on the cheek, arms, nose and ear (Fig. 1). Histopathologically, eight pure and four mixed cases were found, being the median Breslow thickness, 7.3mm, with no significant differences being reported between pure and mixed cases. Only 25% of the cases (3 cases) were pigmented, 50% (6 cases) presented neurotropism, and all cases lymphoid aggregates. Patient staging (computed tomography) turned out negative in all patients at diagnosis except for one patient, 16% (2 cases) showed positive sentinel lymph nodes, all of which were of the mixed subtype, and 25% (3 cases) had local recurrences. A total of 75% (6 cases) of all pure DMs were stage IIB, and 25% (2 cases), stage IV. A total of 50% (2 cases) of all mixed DMs were stage IIB, and the remaining 50% (2 cases), stages III and IV. Systemic therapy with anti-PD-L1 and/or anti-CTLA4 was started in 41% of the patients (5 patients), with excellent responses in all them (Table 1). No patient has died due to melanoma to this date. Twenty-five mutations were found in 13 different genes – 21 were somatic mutations – and four copy number variations with amplifications. The most widely mutated gene was the TP53, found in 50% (6 cases) of all DMs. Mutations in the EGFR, IDH1 and RET genes were the next most widely found in 16% (2 cases) of all DMs reported. Other mutations found were in the ALK, MET, CTNNB1, CD274/PD-L1, FGFR 1 and 3 genes. Only one case of DM exhibited the mutated BRAF gene, while no mutated gene was ever found in three DMs (Table 2). Not differences were found in Breslow thickness or in pure or mixed DM cases with mutations in TP53 vs the remaining cases of DMs.
Clinical and histopathological characteristics of patients diagnosed with DM.
| Case | Age/sex | Clinic | Pigmented | Neurotropism | Lymphoid aggregates | Extension study (CT) | Sentinel lymph node | Local recurrence | Inmunotherapy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 66Male | Scalp nodule | No | No | Yes | Negative | Negative | Yes | Ipilimumab→Nivolumab |
| 2 | 64Male | Scalp nodule | No | No | Yes | Negative | Negative | No | No |
| 3 | 89Female | Cheek plaque | Yes | Yes | Yes | Negative | Negative | No | No |
| 4 | 27Female | Nose nodule | No | Yes | Yes | Negative | Negative | Yes | Nivolumab+Ipilimumab |
| 5 | 95Female | Cheek nodule | No | Yes | Yes | Negative | Negative | No | No |
| 6 | 70Male | Arm nodule | Yes | No | Yes | Negative | Positive | No | Nivolumab |
| 7 | 79Female | Scalp nodule | No | No | Yes | Negative | Positive | Yes | Nivolumab |
| 8 | 92Male | Scalp nodule | No | Yes | Yes | Negative | Negative | No | No |
| 9 | 87Male | Scalp nodule | No | No | Yes | Negative | Negative | No | No |
| 10 | 80Male | Cheek nodule | No | No | Yes | Negative | Negative | No | No |
| 11 | 42Male | Back nodule | Yes | Yes | Yes | Positive | Unrealized | No | Nivolumab |
| 12 | 68Male | Ear nodule | No | Yes | Yes | Negative | Negative | No | No |
Subtype, Breslow thickness and mutations of DM tumors.
| Case | DM type | Breslow (mm) | Mutated gene | Type | Chr | Exon | Mutation: amino acid change | Ratio |
|---|---|---|---|---|---|---|---|---|
| 1 | Pure | 25 | RET | Somatic | 10 | c.2428G>A; p.(Gly810Ser) | ||
| 2 | Pure | 5 | FGFR1 | Somatic | 8 | c.1774G>A; p.(Val592Met) | ||
| RET | Somatic | 10 | c.1894G>A; p.(Glu632Lys) | |||||
| TP53 | Somatic | 17 | 5 | c.637C>T; p.(Arg213Ter) | ||||
| ALK | Copy number variation | 2 | Amplification: 1.29 | |||||
| ERBB2 | Copy number variation | 17 | Amplification: 1.23 | |||||
| 3 | Mixed | 3.7 | TP53 | Somatic | 17 | 8 | c.842A>G; p.(Asp281Gly) | |
| MET | Somatic | 17 | c.2962C>T; p.(Arg988Cys) | |||||
| EGFR | Copy number variation | 7 | Amplification: 4.46 | |||||
| 4 | Pure | 1.5 | None | |||||
| 5 | Pure | 10 | None | |||||
| 6 | Mixed | 7 | BRAF | Somatic | 7 | c.1798_1799delGTinsAA; p.(V600K) | ||
| IDH1 | Somatic | 2 | c.394C>T; p.(R132C) | |||||
| CTNNB1 | Somatic | 3 | c.109T>C; p.(S37P) | |||||
| 7 | Mixed | 8.6 | none | |||||
| 8 | Mixed | 7.3 | FGFR3 | Somatic | 15 | c.1922A>C; p.(Asp641Ala) | ||
| ROS1 | Somatic | 6 | 36 | c.5845C>T; p.(Leu1949Phe) | ||||
| TP53 | Somatic | 17 | 7 | c.722C>T; p.(Ser241Phe) | ||||
| 9 | Pure | 17 | TP53 | Somatic | 17 | 7 | c.733G>A; p.(Gly245Ser) | |
| 10 | Pure | 15 | TP53 | Somatic | 17 | c.809T>G; p.(Phe270Cys) | ||
| TP53 | Somatic | 17 | 8 | c.817C>T; p.(Arg273Cys) | ||||
| EGFR | Somatic | 7 | 8 | c.2305G>A; p.(Val769Met) | ||||
| 11 | Pure | 13 | IDH1 | Somatic | 2 | c.394C>T; p.(Arg132Cys) | ||
| 12 | Mixed | 5.2 | TP53 | Somatic | 17 | 4 | c.395A>T; p.(Lys132Met) | |
| CD274/PD-L1 | Copy number variation | 9 | Amplification: 1.61 | |||||
RET: ret proto-oncogene; FGFR: fibroblast growth factor receptor; TP53: tumor protein p53; ALK: anaplastic lymphoma kinase; ERBB2: erythroblastic oncogene B2; MET: MET proto-oncogene, receptor tyrosine kinase; EGFR: epidermal growth factor receptor; BRAF: B-Raf proto-oncogene, serine/threonine kinase; IDH1: isocitrate dehydrogenase (NADP (+)) 1; CTNNB1: β-catenin 1; ROS1: ROS proto-oncogene 1, receptor tyrosine kinase; CD274/PD-L1: programmed cell death 1 ligand.
The diagnosis of DM can be complicated as they are usually nonspecific lesions. Frequent locations are photoexposed regions with sun damage, which happen to be strongly associated with UV radiation. Lymphoid aggregates and perineural invasion are common histopathological findings in DM,4,5 which is a subtype of melanoma with a high mutation rate. In our study, the most widely mutated gene was the TP53 without correlation with Breslow thickness or the DM subtype. The TP53 mutation is associated with cumulative sun damage. Similar results have been obtained in other studies, where the most common mutation found in DM was TP53. However, in other studies, the most widely identified mutation was in the NF1 gene.6 In our panel of studied genes, the NF1 gene was not included, which accounts for a limitation of our results. In contrast, the BRAF mutation is rare in this subtype of melanoma.7,8 DMs have a good response to immunotherapy (anti PD-L1/anti-CTLA4) and prognosis looks good.9,10
Conflict of interestThe authors declare that they have no conflict of interest.