The identification of B-Raf proto-oncongene (BRAF) mutation and the emergence of targeted therapy marked a turning point in the treatment of melanoma. The study of mutation status concordance between primary tumors and metastases in this cancer has major treatment implications as it facilitates the selection of candidates for targeted therapy. This review analyzes the evidence on the level of mutation status concordance between primary tumors and different types of metastases in cutaneous melanoma and provides an overview of the advantages and disadvantages of the various methods used to detect BRAF mutations.
La mutación en el oncogén BRAF en melanoma y la aparición de terapias dirigidas frente a ella han supuesto un antes y un después en el tratamiento de esta enfermedad. El estudio del estado mutacional en las metástasis y su concordancia con el tumor primario tiene además una gran implicación terapéutica en estos pacientes, pues permite seleccionar candidatos a estos tratamientos. El objetivo de esta revisión es conocer las evidencias disponibles sobre el grado de concordancia en los distintos tipos de metástasis en el melanoma cutáneo, así como las ventajas y desventajas de los distintos métodos de detección de la mutación en BRAF.
Mutation status concordance between primary and metastatic tumors is a critical area of research in oncology that has become even more important with the introduction of therapies that target specific mutations, such as those on the B-Raf proto-oncongene (BRAF).
Concordance has been studied intensively in other malignant tumors but not in melanoma. Most such studies have shown high levels of primary–metastatic concordance: in esophageal cancer, 85% concordance for ERBB2 (formerly HER2) tumors1; in colon cancer, 98% to 100% concordance for BRAF2–5; and also in colon cancer, 92% to 100% concordance for the KRAS proto-oncogene (KRAS).2,3,6–14 Some authors, however, have reported nonnegligible levels of discordance between primary tumors and their corresponding metastases: in esophageal cancer, discordance of 22.5% for ERBB2/HER2,15 and in colon cancer, 32.4% discordance for KRAS.5,16
Few studies have analyzed mutation concordance in melanoma, however, and the results are heterogeneous. Some authors have reported high concordance17,18 while others have reported levels around 50%.19–21 Most studies based their findings on only a single method for identifying the mutation and used small sample sizes, undermining the validity of conclusions. This review summarizes the current state of knowledge concerning BRAF mutation status concordance in cutaneous melanoma, with reference to the various methods used to identify mutations in clinical practice.
BRAF Mutation in MelanomaThe BRAF protein, a key participant in the mitogen-activated protein-kinase (MAPK) pathway, is coded by the BRAF oncogene on chromosome 7 (7q34).22 One of the most common mutations—V600E at the nucleotide position of codon 600 on exon 15—leads to a change of valine to glutamic acid (GTG>GAG) in the protein. V600E occurs in 85% to 90% of cases.22–24 The next most common mutation, V600K (GTG>AAG), causes a valine to lysine change and has been reported to occur in 20% to 21% of cases.25–28 However, the prevalence in some populations is higher.25,28 Other less frequent mutations are V600R (GTG>AGG), V600D (GTG>GAT), V600E2 (GTG>GAA), V600A, V600G, K601N, K601E, L597R, L597Q, G596R, and D594N.29,30
Alterations in the BRAF protein lead to constitutive activation of the MAPK pathway, obviating the need for Ras activation. Mutation increases the protein's kinase activity 10-fold over the activity coded by unmutated, wild-type BRAF (BRAFWT)22,31 and leads to neoplastic cell proliferation by continuous transcription. Somatic alterations in the BRAF oncogene are more common in cutaneous melanomas than in other tumors.22 This mutation, found in 50% to 70% of melanomas, is far more frequent than others, such as neuroblastoma Ras oncogene (NRAS) or p16 and p53 tumor suppressor gene mutations.32,33
Detecting BRAF MutationsSeveral methods are available for detecting BRAF mutations. The choice of one over another is based on ease or provision of rapid results, on sensitivity or specificity, and on cost.
SequencingSanger sequencing and pyrosequencing are the most widely used methods for determining mutation status in research. For many years Sanger sequencing of DNA previously amplified by polymerase chain reaction (PCR) was considered the reference method for detecting acquired mutations in tumors and for sequencing the human genome. This technique can detect base substitutions, deletions, and insertions but is unable to detect alterations in duplicated chromosomes or translocations.30 This type of sequencing requires time (18 to 19hours), special equipment that is not available in many laboratories, and qualified staff to interpret the results. The Sanger method is highly specific, but its main limitation is that sensitivity is low (92.5%),34 obliging a tumor load of up to 5% in samples.30 The high rate of false negatives increases the risk that patients who do in fact carry the mutation might not be treated with BRAF protein inhibitors (BRAFi).
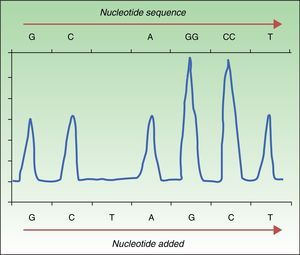
Pyrosequencing, or sequencing by synthesis, relies on detecting the release of pyrophosphate by DNA polymerase on incorporation of the next complementary nucleotide in the DNAss chain. To sequence in this way, DNA is amplified by PCR, followed by hybridization of the DNAss chain with a primer before incubation with enzymes. The strand is washed in sequentially added solutions of A, C, G, and T nucleotides. The pyrophosphate released on incorporation of a nucleotide reacts with the adenosine triphosphate sulfurylase and luciferase enzymes to produce light (chemiluminescence). The light produced is detected and graphed as a peak in a pyrogram (Fig. 1). Intensity, indicated by the height of the peak, reveals whether more than a single nucleotide is present.35,36 This approach detects not only a mutation's presence or absence but also the proportion of DNA carrying the mutation. Pyrosequencing has proven to be much more sensitive than Sanger sequencing,30 and it detects not only the BRAFV600E mutation but also less frequent ones (V600K, V600D, V600R, and K601E).30,34 The technique is rapid (completed in <24hours), but errors are frequent when a nucleotide is sequenced 6 or 7 times (homopolymers). Moreover, whereas Sanger sequencing detects strands up to 800 to 1000 nucleotides long, pyrosequencing is only able to detect strands that are 300 to 500 nucleotides long. The main disadvantage of pyrosequencing is the high cost of reagents and the need for special equipment that is not always available in conventional laboratories.
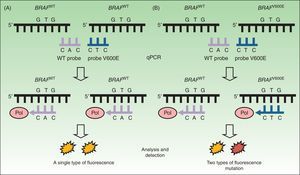
Real-Time PCRDiagnostic tests based on real-time quantitative reverse transcription PCR (RT-qPCR) are now the most widely used in clinical laboratories to detect mutations in tumor samples. This approach is rapid (<8hours) and offers a level of sensitivity (97.5%34,37,38) that is superior to that of the Sanger method. RT-qPCR uses a thermal cycler and fluorescence sensors (Fig. 2) to detect DNA mutations in formalin-fixed, paraffin-embedded tissue. To study the presence or absence of mutations in the BRAF oncogene, a specific sequence on exon 15 is targeted. RT-qPCR is therefore both rapid and economical. Three samples can be analyzed simultaneously without loss of reagent yield, and the technique uses devices already present in the laboratory for detecting other mutations of interest in routine clinical practice (eg, KRAS in colon cancer or the EGFR in nonsmall cell lung cancer).
Real-time quantitative reverse transcription polymerase chain reaction to detect mutations in BRAFV600. A, DNA study of a tumor without the BRAFV600E mutation. Binding with a single probe (for WT) resulted in a single fluorescence signal interpreted to indicate “not mutated.” B, DNA study of a tumor with the BRAFV600E mutation. Binding with 2 probes resulted in emission of 2 types of fluorescence to indicate “mutated.” WT refers to wild-type; Pol, DNA polymerase enzymes; qPCR, quantitative polymerase chain reaction.
Source: Adapted from Curry et al.30
Several devices have recently been approved for RT-qPCR. The Cobas 4800 BRAFV600 Mutation Test (Roche Molecular Systems) has become the most widely used platform since its approval by the US Food and Drug Administration (FDA) for patients who are candidates for treatment with vemurafenib. The clinical usefulness of the Cobas test was validated for identifying tumors carrying the BRAFV600E mutation, which it detected in 100% of samples.39 It also proved useful for other mutations: BRAFV600K (detected in 70% of samples),39BRAFV600D and BRAFV600E2.38,40 The lower limit of detection of the Cobas platform is 4.4% of mutated alleles per 1.25ng/μL. However, other mutations, such as V600K and V600E2, require higher percentages (31% and 68%, respectively).30 Errors are often related to the presence of melanin, an inhibitor of PCR.
When RT-qPCR was used by Yancovitzetal.21 to detect the BRAFV600E mutation in 112 melanoma samples, they found that 75.9% were positive (66.7% of primary tumors and 77.7% of metastases), whereas conventional sequencing found only 32.1% of the cases in the same samples (38.9% in primary tumors and 30.9% in metastases). Moreover, all of the tumor mutations identified by conventional sequencing were found by RT-qPCR.
This technique was also shown to be useful for cytologic analysis of fine-needle aspirate biopsies.41 Agreement between RT-qPCR and Sanger sequencing was 93% in 117 paraffin-embedded biopsies that were obtained for cytology and were composed of at least 50% tumor cells.
Anti-VE1 Monoclonal AntibodyA monoclonal antibody for VE1, the protein expressed by the BRAFV600E mutation, can be detected immunohistochemically in paraffin-embedded material from tumor samples.42–44 Sensitivity for the V600E mutation has been reported to be 100%,34,45 but other mutations (including BRAFV600K) have not been detected. Given that immunohistochemistry is a rapid, economical procedure within the scope of routine practice in hospital pathology laboratories, it promises to be viable for clinical use. However, neither the US FDA or the European Medicines Agency have yet approved it for use in candidates for BRAFi therapy. Moreover, since this therapy has been shown to be effective in treating tumors with the V600K mutation, the exclusive use of immunohistochemistry would not identify BRAFi treatment candidates who have the V600K mutation.
Liquid BiopsySo-called liquid biopsy has been studied in the context of other solid tumors46 and is gaining importance in the evaluation of melanoma. The term encompasses the detection of circulating tumor DNA (ctDNA) and circulating microRNA.47 The use of RT-qPCR in plasma to detect either ctDNA or circulating tumor cells released after apoptosis or necrosis is a promising approach for following patients with metastatic melanoma. The method is currently being validated not only for detecting primary–metastatic mutation discordance but also for evaluating response to treatment and recurrence.
It therefore seems reasonable to combine immunohistochemistry with molecular approaches such as RT-qPCR or pyrosequencing to identify BRAF mutations in patients with metastatic melanoma.
Primary–Metastatic BRAF Mutation Status ConcordanceGiven that primary–metastatic mutation status discordance has been seen in other tumors48 and is a reason for resistance to treatment, this possibility must be studied in cancer patients who are candidates for targeted therapy. Other melanoma mutations (on NRAS, for example) have been shown to have high concordance with the primary tumor.49
Our review of studies of BRAF mutation status concordance in melanoma to date reveals that most have evaluated small samples.19–21 Exceptions are a study by Colombinoetal.17 and a recent one by our group.50 Reviewing the concordance rates in the published studies, we can observe that they show different results (Table 1).
Primary–Metastatic BRAFV600 Mutation Status Concordance in the Literature a
| N, Primary Tumor | N, Metastatis | Lymphatic | Soft-Tissue | Visceral | Method | Discordance | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Conc | Disc | N | Conc | Disc | N | Conc | Disc | |||||
| Riveiro et al.50 | 140 | 171 | 100 | 100 (100%) | 0 | 62 | 62 (100%) | 0 | 9 | 9 (100%) | 0 | PCR+IH+SS | – |
| Colombino et al.17 | 102 | 165 | 84 | 78 (93%) | 6 (7%) | 36 | 29 (81%) | 7 (19%) | 45 | 43 (96%) | 2 (4%) | SS | BRAFM/BRAFWT |
| Saroufim et al.51 | 33 | 33 | 22 | 20 (91%) | 2 (9%) | 7 | 4 (57%) | 3 (43%) | 4 | 2 (50%) | 2 (50%) | PCR+IH | BRAFM/BRAFWT |
| Shinozaki et al.19 | 13 | 13 | 12 | 7 (58%) | 5 (42%) | 1 | 1 (100%) | 0 | SS | BRAFWT | |||
| Lin et al.20 | 2 | 10 | 5 | 2 (40%) | 3 (60%) | 2 | 0 | 2 (100%) | 3 | 1 (33%) | 2 (67%) | SS | BRAFM/BRAFWT |
| Omholt et al.18 | 71 | 51b | 49 (96%) | 2 (4%) | SS | BRAFWT | |||||||
| Houben et al.54 | 24 | 24b | 19 (79%) | 5 (21%) | SS | BRAFM/BRAFWT | |||||||
| Akslen et al.52 | 17 | 17b | 15 (88%) | 2 (12%) | SS | BRAFWT | |||||||
| Yancovitz et al.21 | 18 | 18b | 10 (56%) | 8 (44%) | SS+PCR | BRAFM/BRAFWT | |||||||
| Busam et al.53 | 1 | 1b | 1 (100%) | 0 | SS+IH | – | |||||||
Abbreviations: Conc, concordance; Disc, discordance; IH, immunohistochemistry; PCR, polymerase chain reaction; SS, Sanger sequencing; BRAF, B-Raf proto-oncogene; M (superscript), mutant; WT (superscript), wild-type.
Colombinoetal.17 reported a low rate of the discordant cases (in 15 of 165 cases), 53% of the primary tumors were BRAFWT with mutant BRAF (BRAFM) in metastasis, whereas 47% primary tumors tested BRAFM but had BRAFWT metastases.
Our group observed a higher rate of concordance in a larger multicenter study of 140 primary tumors and 171 metastatic pairs.50 Using the RT-qPCR Cobas® platform we found concordance in 83.6% of the pairs. When we added immunohistochemistry findings for VE1 protein expression and Sanger sequencing, concordance reached 100%.
Other authors studying smaller samples have also reported high rates of concordance. In a study of 71 primary melanomas, Omholtetal.18 were able to evaluate 51 matched metastatic tumors, although they did not specify the type of metastasis. There were only 2 discordant cases, both primary BRAFWT tumors with BRAFM metastases. This study found in 17 consecutive cases of metastasis only a single discordant pair in a patient with a primary BRAFM tumor with both BRAFM and BRAFWT metastases. Saroufimetal.51 studied 40 primary tumors with paired metastases, of which only 33 had adequate material to complete the evaluation. In 6 of the cases, RT-qPCR findings could not be evaluated, so the authors resorted to immunohistochemistry. They confirmed primary–metastatic concordance in 26 of the 33 cases (79%). Five discordant cases were BRAFM primary tumors paired with BRAFWT metastases, and 2 were BRAFWT primary tumors paired with BRAFM metastases. Akslenetal.52 studied 18 cases and were able to report results for 17 pairs. Only 2 pairs, both BRAFWT primary tumors, were discordant. Busametal.53 studied cases in which mutation status results obtained by sequencing were available for comparison with the results of immunohistochemical staining for VE1 protein expression. Among them was a BRAFM primary tumor with concordant metastasis.
However, when even smaller samples were studied, the authors saw higher proportions of discordance. Houbenetal.54 studied NRAS and BRAF status in 24 pairs, finding 7 cases of primary–metastatic discordance. Three were in BRAFWT primary tumors with BRAFM (V600E, V600R, and V600K) metastases. Another 2 cases were in mutant BRAFV600E primary tumors with BRAFWT metastases that acquired NRAS mutations, which had not been present in the primary tumor. The remaining two cases had only NRAS mutation discordance. Yancovitzetal.21 studied concordance in 18 primary tumors with BRAF and their corresponding metastases. The 8 discordant cases were in 6 BRAFWT primary tumors with BRAFM metastases, plus 2 BRAFM primary tumors with BRAFWT metastases. The mutation status of 9 of the primary tumors was heterogeneous, with more than a single cell population within the same tumor, a phenomenon that has been described by other authors.55,56 Shinozakietal.19 detected primary–metastatic discordance in 8 out of 13 cases. All were BRAFWT primary tumors with BRAFM metastases. The authors suggested the mutations might have been acquired during disease progression. Finally, the selection of BRAFM alleles during the progression of initially heterogeneous primary tumors was observed by Linetal.20 in samples from 3 patients. In 1 of the cases, the mutation was found on recurrence of a primary tumor that had not initially been positive for the mutation. In another case, the primary tumor was heterogeneous (BRAFWT plus BRAFV600K) and 7 of the 9 visceral and subcutaneous metastases showed a predominance of BRAFV600K. The third patient had a primary tumor in which BRAFV600K was present, but node metastases contained more of mutated alleles. The authors of this study pointed out that even acral and mucosal melanomas can harbor small BRAFM subpopulations in the primary tumor that can later predominate on metastasis.
Concordance in Subsequent MetastasisSeveral concordance studies have analyzed DNA sequencing data for several metastases in the same patient even though mutation status for the primary tumor was unavailable. In an interesting study in 15 patients, Sigalottietal.57 analyzed 15 initial metastases and 19 subsequent metastases. They found that BRAF mutation status (3 BRAFWT and 12 BRAFM) was maintained in all but 2 cases, in which later metastases from the primary tumors were homozygous although the earlier metastatic tumors had been heterozygous. These observations suggest the stability of established mutation status in metastatic melanoma regardless of location or time frame. Saroufimetal.58 also studied subsequent metastases and Niessneretal.59 studied paired cerebral and extracerebral metastases, finding identical patterns of BRAF, NRAS, and KIT oncogene mutations. In contrast, when Changetal.56 sequenced DNA in frozen tissue in 3 cases of multiple metastases, they found complete mutation status concordance in only a single case. The other 2 were discordant.
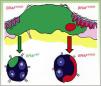
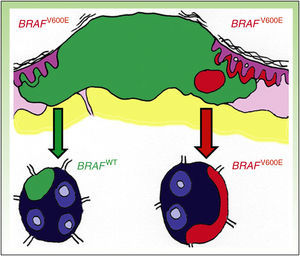
DiscussionDiscordance between primary tumors and their corresponding metastasis can be explained by a theory recently advanced by Yancovitzetal.,21 according to which different subclone populations can coexist in a single tumor (Fig. 3), a status referred to as intratumoral heterogeneity. The presence of different cell populations, some carrying a mutation and others not, allows both BRAFM and BRAFWT metastases to develop. Similarly, resistance to targeted therapy as well as disease progression through clonal selection, which has been observed in some patients who initially responded well to BRAFi therapy, can be explained by both intratumoral and intertumoral heterogeneity (presence of different subclones in primary and metastatic tumors).
The studies in the larger sample sizes suggest that discordance is low or nearly absent in cutaneous melanoma. The findings of discordance in some studies could be attributed to the presence of mutations other than V600E or to the use of a single detection method, as some have low sensitivity. The study with the largest sample size, one which also used the largest number of detection techniques, found 100% concordance,50 supporting the theory that melanoma mutation status stays stable during progression.
In any case, though the prevalence of discordant metastases may be low, the possibility should not be ignored given that patients with discordant tumors who might be candidates for BRAFi therapy may be affected. Not detecting mutation status might deny them the treatment.
It therefore seems reasonable to propose that the ideal tissue for mutation status assessment would come from the most recent metastasis. However, when such tissue is unavailable for analysis, the high levels of concordance found in larger studies suggests that mutations in the primary tumor should be studied instead. Similarly, new metastases that occur when progression takes place during BRAFi therapy should be analyzed for clones that do not carry the same mutation. Moreover, combining techniques—such as RT-qPCR and immunohistochemistry—would increase detection sensitivity.
Ethical DisclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Data confidentialityThe authors declare that no private patient data are disclosed in this article.
Right to privacy and informed consentThe authors declare that no private patient data are disclosed in this article.
Conflicts of InterestThe authors declare that they have no conflicts of interest.
Please cite this article as: Godoy-Gijón E, Yuste-Chaves M, Santos-Briz Á. Concordancia mutacional de BRAF entre melanoma primario cutáneo y sus correspondientes metástasis. Revisión de la evidencia actual. Actas Dermosifiliogr. 2017;108:894–901.