Onychomycosis is known to have predisposing factors and a high prevalence within families that cannot be explained by within-family transmission. We determined the frequency of HLA-B and HLA-DR haplotypes in 25 families of Mexican patients with onychomycosis in order to define the role of the class II major histocompatibility complex (MHC) in genetic susceptibility to this infection. Seventy-eight subjects participated in the study, 47 with onychomycosis and 31 healthy individuals. The frequencies of the HLA-B and HLA-DR haplotypes were compared with those found in first-degree relatives without onychomycosis and in a historic control group of healthy individuals. The frequencies in the controls were similar to those of the healthy relatives of the patients. However, on comparison of the patients with historic controls, we detected a higher frequency of the HLA-DR8 haplotype (P=.03; odds ratio, 1.89; 95% confidence interval, 0.98–36). These findings suggest that there are polymorphisms in genes of the MHC that increase susceptibility to onychomycosis, particularly haplotype HLA-DR8.
En la onicomicosis hay factores predisponentes conocidos y una alta prevalencia en familiares no explicada por transmisión intrafamiliar. Analizamos la frecuencia génica de alelos de los genes HLA-B y HLA-DR en 25 familias de pacientes mexicanos con onicomicosis para conocer el papel del complejo mayor de histocompatibilidad (MHC) clase II en la susceptibilidad genética a esta enfermedad. Se estudiaron 78 individuos, 47 de ellos con onicomicosis y 31 sanos. Se determinaron las frecuencias génicas de los alelos de los loci HLA-B y HLA-DR y se compararon con las presentes en familiares de primer grado sin onicomicosis y con un grupo de individuos sanos. Estas fueron semejantes a las de familiares sanos; sin embargo, al comparar pacientes con controles históricos se encontró un aumento de la frecuencia del alelo HLA-DR8 (p=0,03; OR=1,89; IC 95%: 0,98–36). Estos datos sugieren que dentro del MHC existen genes de susceptibilidad al desarrollo de onicomicosis; en particular con el alelo HLA-DR8.
Onychomycosis is one of the most common nail disorders and is caused by the dermatophyte Trichophyton rubrum in 70% to 85% of cases.1,2 Among the main predisposing factors are humidity, maceration, occlusion, trauma, diabetes, and immunosuppression.1–3 The host's immune response against dermatophyte infections is poorly understood, but it is likely that epidermal Langerhans cells process the metabolites produced by the fungus and present them to neutrophils and lymphocytes. It has, however, been postulated that patients with onychomycosis, apart from having reduced delayed-type hypersensitivity reactions, may not mount a sufficient immune response to eliminate the dermatophyte responsible for the infection.4 This selective, and possibly induced, immune deficiency, may be relatively common, as up to 20% of the general population have chronic dermatophytosis.3,5,6 On the other hand, there have been reports of onychomycosis due to T. rubrum in members of the same family, suggesting the presence of a genetic component.5–8 One group of authors, using pedigree analysis, even showed an autosomal dominant pattern of inheritance.7 The presence of onychomycosis in members of the same family cannot be explained by intrafamilial transmission as the prevalence of this disease is very low in people who are married to or have been living with an infected person for years.6,7 In any case, depressed cellular immunity is a known risk factor for onychomycosis, and intact cellular immunity is critical for eliminating dermatophytes from the skin. The highly polymorphic major histocompatibility complex (MHC) genes are involved in regulating the immune response and are therefore useful for identifying genes associated with susceptibility to certain diseases and, in particular, infections. Thus, the polymorphic human leukocyte antigen (HLA) genes code for molecules involved in the inflammatory response (e.g., components of the complement system and the tumor necrosis factor),4,9,10 and they also exhibit a strong linkage disequilibrium. Consequently, any allele associated with a particular disease may be a marker of susceptibility to that disease, but it must be determined which is the primary susceptibility allele and which is indirectly associated with the disease.11–13 HLA-DR4 is the most common allele (23%) in mestizo Mexicans,9,10,13 and open-population studies have found alleles that confer resistance to onychomycosis in this population.14 We have designed a family-based study to expand current knowledge about the genetic component involved in susceptibility to onychomycosis.
Case DescriptionsA segregation analysis of families in which at least 1 member had onychomycosis was performed by the dermatology department of Hospital General Dr. Manuel Gea González in Mexico DF, Mexico, between January and July 2010. The study design and methods complied with the principles of the Declaration of Helsinki. We included 47 mestizo Mexicans with a clinical diagnosis of onychomycosis confirmed by direct examination and/or by a positive dermatophyte culture. Twenty-five of the patients were blood relatives. We also included a healthy control group consisting of 31 first-degree blood and non-blood relatives of the patients. Once informed consent had been obtained, genomic DNA was extracted from peripheral blood mononuclear leukocytes for subsequent amplification of specific regions of the HLA-B and HLA-DR loci by polymerase chain reaction at the transplant department of the National Institute of Medical Sciences and Nutrition Salvador Zubirán. Alleles at each locus were identified by low-resolution HLA typing using sequence-specific oligonucleotide probes. Statistical analyses were performed using the StatCalc program in the Epi Info software package.
We compared HLA-B and HLA-DR haplotype frequencies with those of a historical control group consisting of 381 unrelated healthy individuals with 762 distinct haplotypes.13 All of the people in the control groups were born in Mexico, as were their parents and grandparents. They were part of a healthy cohort of organ donors and had no history of autoimmune or HLA-related disease. The prevalence of onychomycosis in this group is unknown.
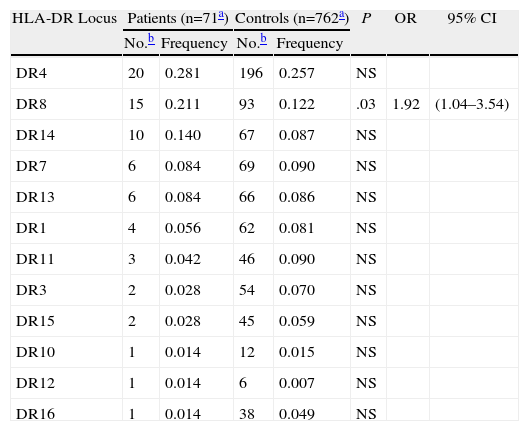
Blood samples were obtained from 78 individuals (47 patients and 31 healthy relatives) from 25 families. HLA-B and HLA-DR haplotype frequencies were calculated for both patients and controls, but the very small number of haplotypes (n=28) in the control group made it impossible to compare the 2 groups. Given the high diversity of HLA-B haplotypes in relation to HLA-DR haplotypes, we decided to limit the comparison to alleles at the HLA-DR locus, which showed a clearer pattern of clustering. Table 1 shows the allele frequencies for individuals with onychomycosis and healthy individuals from the same ethnic group. The most common allele in both groups is HLA-DR4, possibly suggesting that both groups have the same ethnic origin. Most other allele frequencies were similar between patients and controls, with the exception of HLA-DR8, which was significantly more frequent in the patient group than in the control group (P=.03; OR, 1.92; 95% CI, 1.04–3.54).
Frequency of Alleles at the HLA-DR Locus in Patients With Onychomycosis and Healthy Controls.
| HLA-DR Locus | Patients (n=71a) | Controls (n=762a) | P | OR | 95% CI | ||
| No.b | Frequency | No.b | Frequency | ||||
| DR4 | 20 | 0.281 | 196 | 0.257 | NS | ||
| DR8 | 15 | 0.211 | 93 | 0.122 | .03 | 1.92 | (1.04–3.54) |
| DR14 | 10 | 0.140 | 67 | 0.087 | NS | ||
| DR7 | 6 | 0.084 | 69 | 0.090 | NS | ||
| DR13 | 6 | 0.084 | 66 | 0.086 | NS | ||
| DR1 | 4 | 0.056 | 62 | 0.081 | NS | ||
| DR11 | 3 | 0.042 | 46 | 0.090 | NS | ||
| DR3 | 2 | 0.028 | 54 | 0.070 | NS | ||
| DR15 | 2 | 0.028 | 45 | 0.059 | NS | ||
| DR10 | 1 | 0.014 | 12 | 0.015 | NS | ||
| DR12 | 1 | 0.014 | 6 | 0.007 | NS | ||
| DR16 | 1 | 0.014 | 38 | 0.049 | NS | ||
Abbreviations: HLA, human leukocyte antigen, OR, odds ratio.
We have identified an allele associated with susceptibility to developing onychomycosis in mestizo Mexicans at the HLA-DR locus, and in particular at HLA-DR8. Interestingly, the frequencies of all of the other alleles at both the HLA-DR and HLA-B loci were similar in individuals with onychomycosis and healthy controls. These findings differ from the results of a recent study by Asz-Sigall et al.,14 in which a significantly lower frequency of HLA-DR6 was found in a similar group of patients compared to controls, suggesting that this allele confers protection against developing onychomycosis. These differences are probably due to variations in the study designs, as Asz-Sigall et al. researched families with multiple cases of the disease. Our findings also differ from those of Ahmed et al.,15 who found a high frequency of HLA-A26, HLA-AW33, and HLA-DR4 in a group of 29 patients with chronic dermatophyte infections. As would be expected, our findings also differ from those reported in 2 studies of Ashkenazi Jews,4,5 a genetically homogenous population with a well-characterized HLA profile. While Zaitz et al.5 reported that HLA-DR53 exerted a protective effect in susceptibility to developing chronic onychomycosis due to T. rubrum, Sadahiro et al.4 found an association between HLA-B14 and resistance to infection, as well as a possible association between HLA-DQB1*06 and susceptibility to chronic dermatophytosis. Nonetheless, these studies agree that these antigens play a critical role in immune response modulation and in the development of chronic fungal infections, such as onychomycosis. The mechanism by which HLA-DR8 increases susceptibility to onychomycosis is complex and probably involves multiple factors, including a genetic component. However, our study provides particularly useful insights, since we included non-blood relatives of the patients with onychomycosis and found that the role of HLA-DR8 was maintained. This genetic factor is polygenic, and the genes analyzed here regulate immune responses and, in particular, adaptive responses. It is important thus that future studies include molecular markers of innate immune responses such as Toll-like receptors on phagocytes.
ConclusionsWe have confirmed that the HLA system plays a role in the T-cell immune response against fungal antigens, and have identified the HLA-DR8 allele to be a likely susceptibility factor for development of onychomycosis in a group of mestizo Mexicans. While we did not identify an HLA allele that conferred resistance to onychomycosis in our group of patients, we should note that the number of HLA haplotypes in healthy individuals was very low.
In future studies analyzing gene frequencies, it would be of interest to increase the number of families in order to obtain a sufficient number of healthy individuals for the control group and not have to resort to a historical control group.
Conflict of InterestsThe authors declare that they have no conflicts of interest.
We are grateful to the Fundación Nacional para la Enseñanza y la investigación and the Fundación Mexicana para la Dermatología, A.C. for providing the funds required for the DNA and HLA analyses in this study.
Please cite this article as: García-Romero M. T, Granados J, Vega-Memije ME, Arenas R. Análisis del polimorfismo genético de los loci HLA-B Y HLA-DR en pacientes con onicomicosis dermatofítica y familiares en primer grado. Actas Dermosifiliogr. 2012;103:59–62.