The influence of human papillomavirus (HPV) on the development of nonmelanoma skin cancer (NMSC) is a topic of debate. HPV types from the beta genus (HPV-β) have been most frequently associated with the development of skin cancer.
ObjectivesTo analyze the prevalence and range of HPV types in NMSC lesions and healthy perilesional skin in immunodepressed and immunocompetent patients and to evaluate the influence of various clinical factors on the prevalence of HPV in skin cancer.
MethodsNested polymerase chain reaction and sequencing were used to detect HPV in 120 NMSC samples obtained by biopsy from 30 kidney transplant recipients and 30 immunocompetent patients. In all cases, a sample was taken from the tumor site and the surrounding healthy skin. Potential confounders were assessed and the data analyzed by multivariate logistic regression.
ResultsHPV DNA was detected in 44 (73.3%) of the 60 samples from immunodepressed patients and in 32 (53.3%) of the 60 samples from immunocompetent patients (adjusted odds ratio, 3.4; 95% CI, 1.2-9.6). In both groups of patients, HPV was more common in healthy perilesional skin than in lesional skin. HPV-β was the most common type isolated.
ConclusionWe found a wide range of HPV types (mostly HPV-β) in the skin of kidney transplant recipients and immunocompetent patients with skin cancer.
La influencia del virus del papiloma humano (VPH) en el desarrollo de carcinoma cutáneo no melanoma es un tema controvertido. VPH-β es el género más frecuente relacionado con el desarrollo de cáncer de piel.
ObjetivosAnalizar la prevalencia y espectro de los tipos de VPH presentes en piel tumoral y piel sana perilesional en pacientes inmunodeprimidos y pacientes inmunocompetentes, así como evaluar la influencia de diferentes factores clínicos en la prevalencia del VPH en cáncer de piel.
MétodosSe determinó la presencia de VPH en 120 muestras mediante PCR nested y posterior secuenciación. Se tomó biopsia de piel de 30 pacientes trasplantados renales y de 30 pacientes inmunocompetentes con cáncer cutáneo tanto de zona tumoral como de piel sana perilesional. Se registraron las variables potenciales de confusión. Los datos fueron analizados utilizando análisis de regresión logística multivariado.
ResultadosADN del VPH fue detectado en 44/60 (73,3%) de las muestras de pacientes inmunodeprimidos y en 32/60 (53,3%) de las muestras de pacientes inmunocompetentes (OR ajustada 3,4 [1,2-9,6]). Al comparar la presencia de VPH en los 2 grupos entre piel tumoral y piel sana perilesional la presencia de VPH en piel sana perilesional fue mayor que en piel tumoral. El género más frecuente aislado fue el VPH-β.
ConclusiónUn amplio espectro de tipos de VPH, la mayoría del género β, se encuentran en la piel de pacientes trasplantados e inmunocompetentes con cáncer cutáneo.
The human papillomavirus (HPV) is a known risk factor for developing cervical cancer.1,2 It is also implicated in the pathogenesis of squamous cell carcinoma of the mucous membranes (vulva, penis, anus) and of the head and neck (pharynx).3 However, its role in the development of nonmelanoma skin cancer (NMSC) is more controversial. Studies of the role of HPV in cutaneous oncogenesis vary widely, in part because of differences in the types of sample analyzed and the methods used for the detection and typing of HPV. Moreover, many studies fail to account for potential confounders that influence the risk of developing skin cancer, particularly factors related to sun exposure.
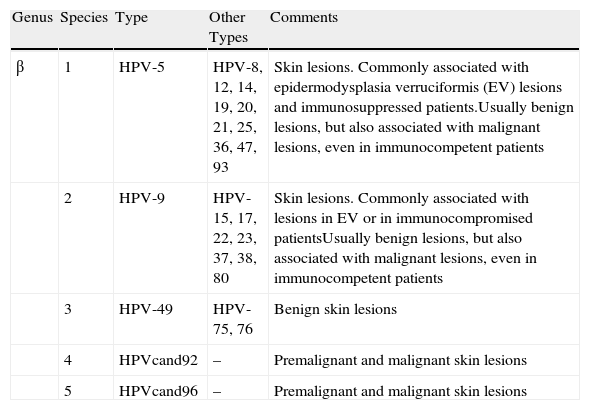
According to current taxonomic classification4 the Papillomaviridae family contains 16 genera, which are identified by Greek letters (α, β, γ, etc.). Each genus contains one or more species, which are numbered, and each species in turn contains types, subtypes, and variants. The classification of the β-papillomavirus (β-PV) genus is shown in Table 1. The relationship between HPV and skin cancer is based on several observations, including the epidemiological association between warts and cutaneous squamous cell carcinoma (SCC) in patients with epidermodysplasia verruciformis,5 the coexistence of epithelial dysplasia and histological signs of viral infection in biopsy,6,7 and the identification of HPV DNA in many NMSCs.8,9 Initially, the mechanisms identified in mucosal oncogenic HPVs were extrapolated in an attempt to identify the molecular strategies or mechanisms by which the β and γ genera induce skin cancer. However, distinct and much more complex mechanisms of action were identified for cutaneous HPV types, possibly involving other cofactors such as ultraviolet (UV) radiation and immunosuppression. Molecular studies performed in cell culture indicate that β-PVs mainly target antiapoptotic mechanisms and block pathways that maintain genomic integrity by repairing damage caused by UV radiation. HPV-8 and HPV-20 of the β genus block the action of the proapoptotic proteins BAK and BAX,10 whose action is similar to but independent of p53. XRCC1, another protein that repairs thymidine dimers induced by UV radiation, can also be intercepted by some forms of HPV, such as HPV-8.11 The HPV proteins E6 and E7 are oncoproteins encoded by the virus. Recent studies have investigated the role of these proteins in the transformation to SCC in transplant patients. E6/E7 transcripts of HPV-8, HPV-9, and HPV-15 have been found in actinic keratoses and SCC, suggesting an active role of HPV in the pathogenesis of these conditions.12 Furthermore, HPV-8 and HPV-5 exert an inhibitory effect on interleukin-8, which is involved in the stimulation of an immune response that targets cells damaged by UV radiation.13
Taxonomic Classification of Genus β Human Papillomavirus Viruses (HPV).
| Genus | Species | Type | Other Types | Comments |
| β | 1 | HPV-5 | HPV-8, 12, 14, 19, 20, 21, 25, 36, 47, 93 | Skin lesions. Commonly associated with epidermodysplasia verruciformis (EV) lesions and immunosuppressed patients.Usually benign lesions, but also associated with malignant lesions, even in immunocompetent patients |
| 2 | HPV-9 | HPV-15, 17, 22, 23, 37, 38, 80 | Skin lesions. Commonly associated with lesions in EV or in immunocompromised patientsUsually benign lesions, but also associated with malignant lesions, even in immunocompetent patients | |
| 3 | HPV-49 | HPV-75, 76 | Benign skin lesions | |
| 4 | HPVcand92 | – | Premalignant and malignant skin lesions | |
| 5 | HPVcand96 | – | Premalignant and malignant skin lesions |
However, given the lack of consistent results demonstrating the presence of HPV DNA in skin cancer, the frequent detection of HPV in normal skin, and the failure to identify a group of skin cancer patients at high risk of developing HPV, the role of HPV in the development of NMSC is unclear.
In the present study we sought to determine the prevalence and spectrum of HPV types in skin tumors and healthy perilesional skin from renal transplant recipients and immunocompetent patients, and to evaluate the influence of different clinical factors on the prevalence of HPV in skin cancer.
MethodsPatients and Study DesignWe determined the presence of HPV DNA in samples of lesional and healthy perilesional skin from immunocompromised and immunocompetent patients. We studied 60 patients (30 renal transplant recipients and 30 immunocompetent patients), who were referred from the Nephrology department to the Dermatology department at the University Hospital Doctor Peset, Valencia, Spain, between January 2010 and January 2012 for evaluation of suspected of NMSC lesions. The study was approved by the hospital's Ethics Committee. Informed consent was obtained from all patients.
Patients were interviewed using a structured questionnaire to collect information on potential confounding variables: age, sex, skin type (Fitzpatrick scale), occupational sun exposure (considered high for patients who spend or had spent most of their working day outdoors), and current and past history of warts and actinic keratosis.
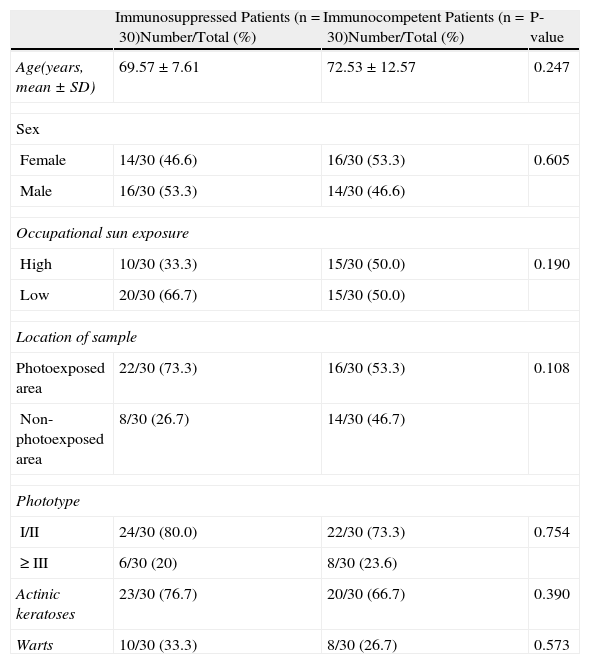
Patient characteristics are shown in Table 2.
Characteristics of Immunocompromised and Immunocompetent Patients.
| Immunosuppressed Patients (n=30)Number/Total (%) | Immunocompetent Patients (n=30)Number/Total (%) | P-value | |
| Age(years, mean±SD) | 69.57±7.61 | 72.53±12.57 | 0.247 |
| Sex | |||
| Female | 14/30 (46.6) | 16/30 (53.3) | 0.605 |
| Male | 16/30 (53.3) | 14/30 (46.6) | |
| Occupational sun exposure | |||
| High | 10/30 (33.3) | 15/30 (50.0) | 0.190 |
| Low | 20/30 (66.7) | 15/30 (50.0) | |
| Location of sample | |||
| Photoexposed area | 22/30 (73.3) | 16/30 (53.3) | 0.108 |
| Non-photoexposed area | 8/30 (26.7) | 14/30 (46.7) | |
| Phototype | |||
| I/II | 24/30 (80.0) | 22/30 (73.3) | 0.754 |
| ≥III | 6/30 (20) | 8/30 (23.6) | |
| Actinic keratoses | 23/30 (76.7) | 20/30 (66.7) | 0.390 |
| Warts | 10/30 (33.3) | 8/30 (26.7) | 0.573 |
Abbreviation: SD, standard deviation.
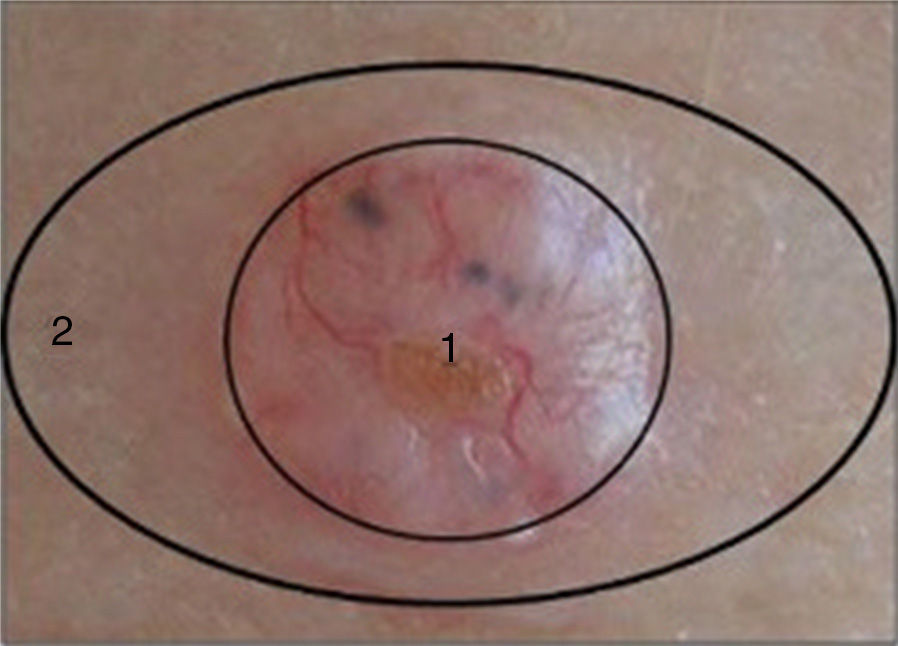
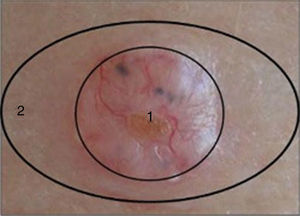
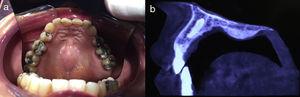
Two samples were taken from each patient as follows: after surgical excision of the suspicious lesion a 3-mm punch was used to take a sample from the center of the lesion (tumor sample) and another from the uninjured portion of the same piece of excised tissue (healthy perilesional skin) (Fig. 1). The samples were stored at -80°C in individual cryotubes until processing. The remainder of the excised tissue was fixed in 10% formalin and embedded in paraffin for histopathological diagnosis. A total of 120 samples were obtained: 60 from transplant recipients and 60 from immunocompetent patients (ie, 30 tumor samples and 30 healthy perilesional skin samples from each group).
Each group of 30 tumor samples collected from immunocompromised and immunocompetent patients consisted of 7 SCC, 10 SCC in situ, and 13 basal cell carcinomas (BCC).
Skin samples were defined as being from exposed areas when they were taken from the face, the neck or nape of the neck, the chest, the upper back, the dorsum of the forearm, and the shins.
Detection of Human PapillomavirusDNA extraction and HPV typing was performed as previously described.14 DNA was extracted from samples using a Maxwell 16 FFPE Tissue LEV DNA purification kit. Different HPV types were identified by nested polymerase chain reaction (PCR) followed by sequencing. DNA amplification was achieved using 100 ng of DNA in a final volume of 25μL (for the 2 rounds of amplification) containing 3.5mM MgCl2, 10mM Tris-HCL, 50mM KCl, 0.1% Triton X-100, 0.2mM of each deoxynucleotide triphosphate, 0.75μM of each primer, 5% dimethyl sulfoxide (DMSO), and 1.25 U Taq DNA polymerase (Promega). A volume of 2.5μL was transferred from the first to the second amplification. The first round of amplification consisted of 45 cycles of denaturation (1.5min at 94°C), annealing (1.5min at 50°C), and extension (1.5min at 72°C). The second round of amplification consisted of 30 cycles of denaturing (1.5min at 94°C), annealing (1.5min at 50°C), and extension (1.5min at 72°C). The primers used in the first round of amplification were FAP64 and FAP59, which have been previously described,15 with positions corresponding to nucleotides 5981-6001 (sense primer, FAP59) and 6458-6436 (antisense primer, FAP64). The primers used in the second round of amplification were FAP6085F and FAP6319R,16 with positions corresponding to nucleotides 6085-6104 (sense primer, FAP6085F) and 6319-6296 (antisense primer, FAP6319R). The position of these primers corresponds to the HPV-8 genome. The amplified 235-base pair product was detected on 3% agarose gel and was purified on a Purelink column (Invitrogen). The sequencing reaction was performed in a final volume of 10μL using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions. The amplification profile used was as follows: 25 cycles of 10seconds at 94°C, 10minutes at 50°C, and 2minutes at 60°C. On completion of the sequencing reaction, the product was purified in alcohol (100% and 70% ethanol). The final product was resuspended in deionized formamide, sequenced using an ABI PRISM ® 310 Genetic Analyzer (Applied Biosystems), and the sequence searched against the NCBI database using the BLASTn algorithm.
Statistical AnalysisCategorical variables are presented as frequencies and percentages and continuous variables as the mean and standard deviation. Differences in the distribution of the variables between groups were evaluated using the χ2 test or the Fisher exact test for categorical variables and analysis of variance for continuous data. The odds ratio (OR) and 95% CI were calculated. The OR, adjusted for occupational sun exposure, skin type, and tumor location, was also estimated. Multiple logistic regression analysis was used to identify variables independently associated with the presence of HPV in skin samples. Analyses were performed using the SPSS statistical analysis package (version 19). Statistical significance was set at P<.05.
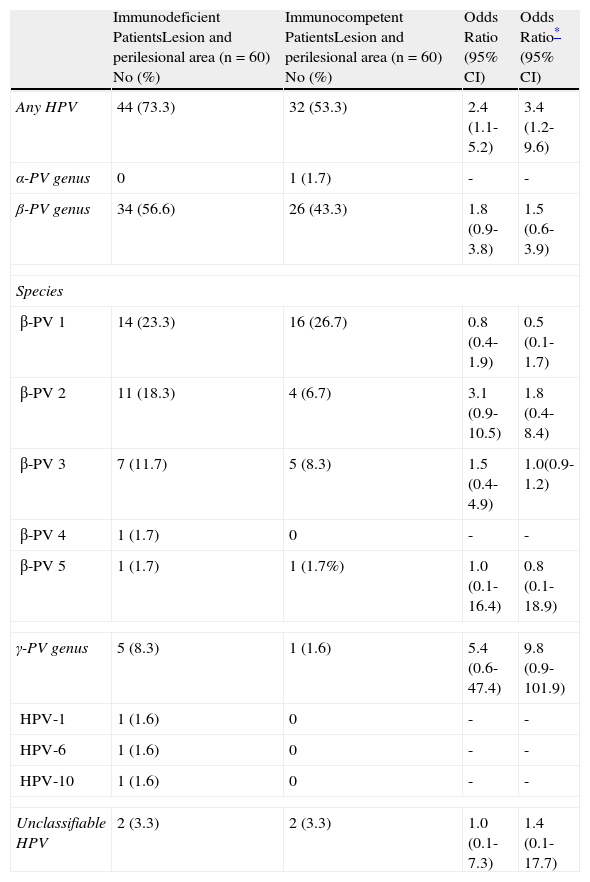
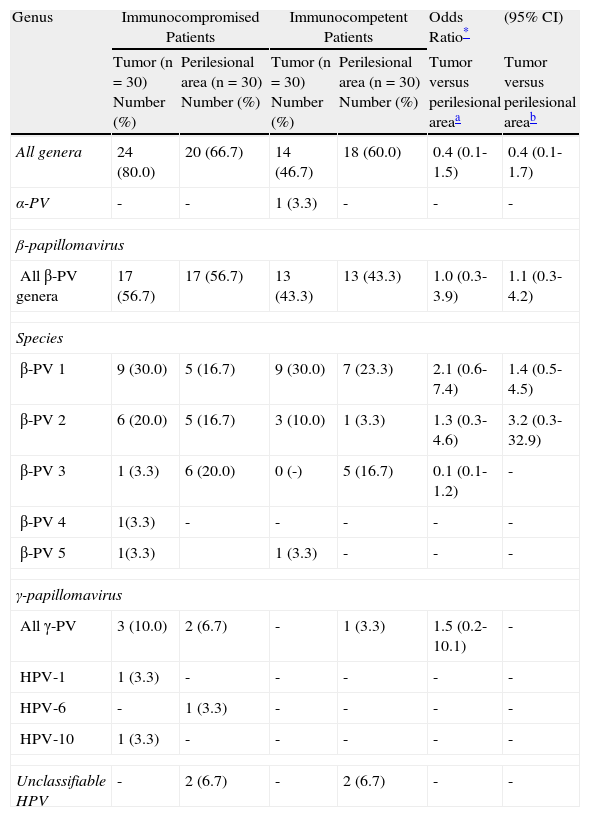
ResultsHuman Papillomavirus in Immunocompetent and Immunocompromised PatientsHPV DNA was detected more frequently in immunocompromised than immunocompetent patients. HPV was detected in 44 of the 60 samples (73.3%) from transplant recipients as compared with 32 of the 60 samples (53.3%) from immunocompetent patients. The difference between groups was significant (adjusted OR, 3.4; 95% CI, 1.2-9.6). Comparison of the different HPV types revealed that the most prevalent form was the β-PV genus, although the relative prevalence of HPV types did not differ significantly between immunocompromised and immunocompetent patients (Table 3).
Distribution of Human Papillomavirus (HPV) Infection in Immunocompromised and Immunocompetent Patients.
| Immunodeficient PatientsLesion and perilesional area (n=60) No (%) | Immunocompetent PatientsLesion and perilesional area (n=60) No (%) | Odds Ratio (95% CI) | Odds Ratio* (95% CI) | |
| Any HPV | 44 (73.3) | 32 (53.3) | 2.4 (1.1-5.2) | 3.4 (1.2-9.6) |
| α-PV genus | 0 | 1 (1.7) | - | - |
| β-PV genus | 34 (56.6) | 26 (43.3) | 1.8 (0.9-3.8) | 1.5 (0.6-3.9) |
| Species | ||||
| β-PV 1 | 14 (23.3) | 16 (26.7) | 0.8 (0.4-1.9) | 0.5 (0.1-1.7) |
| β-PV 2 | 11 (18.3) | 4 (6.7) | 3.1 (0.9-10.5) | 1.8 (0.4-8.4) |
| β-PV 3 | 7 (11.7) | 5 (8.3) | 1.5 (0.4-4.9) | 1.0(0.9-1.2) |
| β-PV 4 | 1 (1.7) | 0 | - | - |
| β-PV 5 | 1 (1.7) | 1 (1.7%) | 1.0 (0.1-16.4) | 0.8 (0.1-18.9) |
| γ-PV genus | 5 (8.3) | 1 (1.6) | 5.4 (0.6-47.4) | 9.8 (0.9-101.9) |
| HPV-1 | 1 (1.6) | 0 | - | - |
| HPV-6 | 1 (1.6) | 0 | - | - |
| HPV-10 | 1 (1.6) | 0 | - | - |
| Unclassifiable HPV | 2 (3.3) | 2 (3.3) | 1.0 (0.1-7.3) | 1.4 (0.1-17.7) |
HPV DNA was detected more frequently in healthy perilesional skin than in lesional skin in both immunocompromised and immunocompetent patients. Comparison between lesional and healthy perilesional skin samples of the prevalence of different HPV genera revealed that the β-PV genus, and in particular β-PV species 1, predominated in both groups and no differences were observed between groups.
Table 4 shows the rates of detection of HPV DNA and the adjusted OR value for skin tumor and perilesional skin samples from immunocompromised and immunocompetent patients. We adjusted the OR according to phototype, sun exposure, and sample location.
Detection of Human Papilloma Virus (HPV) DNA in Lesional and Healthy Perilesional Skin from Immunocompromised and Immunocompetent Patients.
| Genus | Immunocompromised Patients | Immunocompetent Patients | Odds Ratio* | (95% CI) | ||
| Tumor (n=30) Number (%) | Perilesional area (n=30) Number (%) | Tumor (n=30) Number (%) | Perilesional area (n=30) Number (%) | Tumor versus perilesional areaa | Tumor versus perilesional areab | |
| All genera | 24 (80.0) | 20 (66.7) | 14 (46.7) | 18 (60.0) | 0.4 (0.1-1.5) | 0.4 (0.1-1.7) |
| α-PV | - | - | 1 (3.3) | - | - | - |
| β-papillomavirus | ||||||
| All β-PV genera | 17 (56.7) | 17 (56.7) | 13 (43.3) | 13 (43.3) | 1.0 (0.3-3.9) | 1.1 (0.3-4.2) |
| Species | ||||||
| β-PV 1 | 9 (30.0) | 5 (16.7) | 9 (30.0) | 7 (23.3) | 2.1 (0.6-7.4) | 1.4 (0.5-4.5) |
| β-PV 2 | 6 (20.0) | 5 (16.7) | 3 (10.0) | 1 (3.3) | 1.3 (0.3-4.6) | 3.2 (0.3-32.9) |
| β-PV 3 | 1 (3.3) | 6 (20.0) | 0 (-) | 5 (16.7) | 0.1 (0.1-1.2) | - |
| β-PV 4 | 1(3.3) | - | - | - | - | - |
| β-PV 5 | 1(3.3) | 1 (3.3) | - | - | - | |
| γ-papillomavirus | ||||||
| All γ-PV | 3 (10.0) | 2 (6.7) | - | 1 (3.3) | 1.5 (0.2-10.1) | - |
| HPV-1 | 1 (3.3) | - | - | - | - | - |
| HPV-6 | - | 1 (3.3) | - | - | - | - |
| HPV-10 | 1 (3.3) | - | - | - | - | - |
| Unclassifiable HPV | - | 2 (6.7) | - | 2 (6.7) | - | - |
HPV types belonging to the β-PV genus were the most common and were found in 34/60 (56.5%) of the samples from immunocompromised patients and in 26/60 (43.3%) of the samples from immunocompetent patients. Within the β-PV genus species 1 was the most prevalent, and HPV-20 the most common type; no significant differences were detected between groups. The α-PV genus was detected in only 1 sample (in an immunocompetent patient). The γ-PV genus was strongly associated with immunocompromised patients, although this effect was not statistically significant. In 6 instances PCR for HPV was weakly positive, but sequencing could not be performed due to the limited amount of viral DNA available. Four samples of HPV that were not classified in Genbank were detected (Table 4).
Mixed infections (involving multiple HPV types) were detected in 29 of the 120 samples tested, and were more common in immunocompromised (17/60) than immunocompetent (12/60) patients.
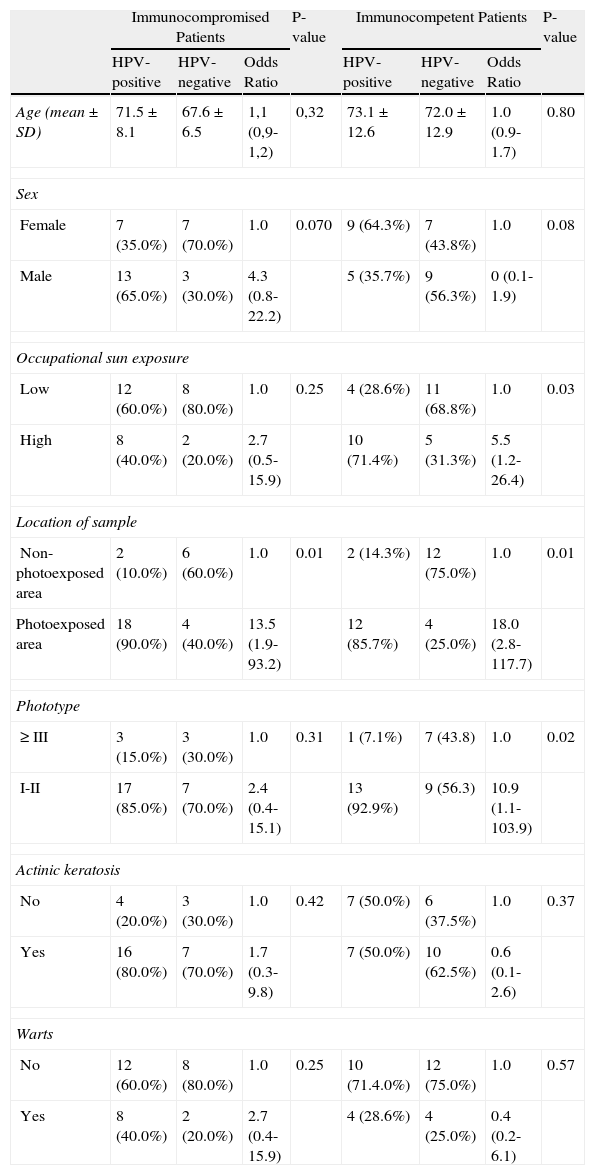
Risk FactorsThe age profile and male to female ratio were similar in the immunocompromised and immunocompetent groups. No significant differences in sun exposure, tumor location, phototype, or history of warts or actinic keratosis were observed between groups. The location of the tumor in an exposed area was the only variable that was significantly associated with an increased risk of HPV in immunocompromised patients. In the immunocompetent group a high level of occupational exposure, tumor location in an exposed area, and a low phototype were significantly more common in HPV-positive patients. In immunocompromised patients the risk of HPV positivity was higher in patients with actinic keratoses and warts, although this difference was not statistically significant (Table 5).
Risk Factors for Human Papillomavirus (HPV) in Immunocompromised and Immunocompetent Patients.
| Immunocompromised Patients | P-value | Immunocompetent Patients | P-value | |||||
| HPV-positive | HPV-negative | Odds Ratio | HPV-positive | HPV-negative | Odds Ratio | |||
| Age (mean±SD) | 71.5±8.1 | 67.6±6.5 | 1,1 (0,9-1,2) | 0,32 | 73.1±12.6 | 72.0±12.9 | 1.0 (0.9-1.7) | 0.80 |
| Sex | ||||||||
| Female | 7 (35.0%) | 7 (70.0%) | 1.0 | 0.070 | 9 (64.3%) | 7 (43.8%) | 1.0 | 0.08 |
| Male | 13 (65.0%) | 3 (30.0%) | 4.3 (0.8-22.2) | 5 (35.7%) | 9 (56.3%) | 0 (0.1-1.9) | ||
| Occupational sun exposure | ||||||||
| Low | 12 (60.0%) | 8 (80.0%) | 1.0 | 0.25 | 4 (28.6%) | 11 (68.8%) | 1.0 | 0.03 |
| High | 8 (40.0%) | 2 (20.0%) | 2.7 (0.5-15.9) | 10 (71.4%) | 5 (31.3%) | 5.5 (1.2-26.4) | ||
| Location of sample | ||||||||
| Non-photoexposed area | 2 (10.0%) | 6 (60.0%) | 1.0 | 0.01 | 2 (14.3%) | 12 (75.0%) | 1.0 | 0.01 |
| Photoexposed area | 18 (90.0%) | 4 (40.0%) | 13.5 (1.9-93.2) | 12 (85.7%) | 4 (25.0%) | 18.0 (2.8-117.7) | ||
| Phototype | ||||||||
| ≥III | 3 (15.0%) | 3 (30.0%) | 1.0 | 0.31 | 1 (7.1%) | 7 (43.8) | 1.0 | 0.02 |
| I-II | 17 (85.0%) | 7 (70.0%) | 2.4 (0.4-15.1) | 13 (92.9%) | 9 (56.3) | 10.9 (1.1-103.9) | ||
| Actinic keratosis | ||||||||
| No | 4 (20.0%) | 3 (30.0%) | 1.0 | 0.42 | 7 (50.0%) | 6 (37.5%) | 1.0 | 0.37 |
| Yes | 16 (80.0%) | 7 (70.0%) | 1.7 (0.3-9.8) | 7 (50.0%) | 10 (62.5%) | 0.6 (0.1-2.6) | ||
| Warts | ||||||||
| No | 12 (60.0%) | 8 (80.0%) | 1.0 | 0.25 | 10 (71.4.0%) | 12 (75.0%) | 1.0 | 0.57 |
| Yes | 8 (40.0%) | 2 (20.0%) | 2.7 (0.4-15.9) | 4 (28.6%) | 4 (25.0%) | 0.4 (0.2-6.1) | ||
Abbreviation: SD, standard deviation.
HPV-positive and HPV-negative indicates the presence and absence of HPV, respectively, in the total number of samples (tumors and perilesional area).
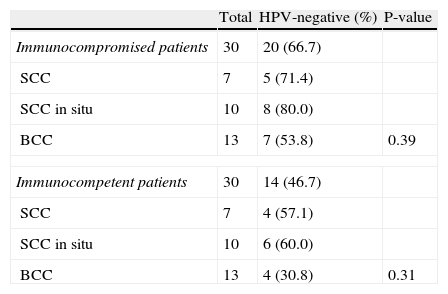
We collected samples from 60 skin lesions: 30 from immunocompromised patients (7 SCC, 10 SCC in situ, and 13 BCC) and 30 from immunocompetent patients (7 SCC, 10 SCC in situ, and 13 BCC). The presence of HPV was more common in SCC in situ in both immunocompromised and immunocompetent patients; HPV DNA was detected in 8/10 (80%) SCC in situ samples from immunocompromised patients and in 6/10 (60%) SCC in situ samples from immunocompetent patients. HPV prevalence was lowest in BCC samples from both immunocompromised and immunocompetent patients, although this effect was not statistically significant (Table 6).
Prevalence of Human Papillomavirus (HPV) in Lesion Samples.
| Total | HPV-negative (%) | P-value | |
| Immunocompromised patients | 30 | 20 (66.7) | |
| SCC | 7 | 5 (71.4) | |
| SCC in situ | 10 | 8 (80.0) | |
| BCC | 13 | 7 (53.8) | 0.39 |
| Immunocompetent patients | 30 | 14 (46.7) | |
| SCC | 7 | 4 (57.1) | |
| SCC in situ | 10 | 6 (60.0) | |
| BCC | 13 | 4 (30.8) | 0.31 |
Abbreviations: BCC, basal cell carcinoma; SCC, squamous cell carcinoma.
In this study we examined a population of immunocompromised and immunocompetent patients to determine the prevalence and spectrum of HPV types present in samples of tumors and healthy perilesional skin. The higher prevalence of HPV found in immunocompetent versus immunocompromised patients is consistent with previously published findings. A study by Harwood and coworkers17 used degenerate PCR to compare HPV prevalence in 148 samples, 85 from immunocompromised patients (44 SCC, 24 BCC and 17 premalignant lesions [ie, actinic keratosis or SCC in situ]) and 63 from immunocompetent patients (22 SCC, 30 BCC, and 11 premalignant lesions). Those authors found a higher prevalence of HPV in the lesions of immunosuppressed versus immunocompetent patients (82% versus 36%, P < .001).
Our analysis revealed a higher prevalence of HPV in healthy perilesional skin than in lesional skin in both immunocompromised and immunocompetent patients, although this effect was not statistically significant. The high prevalence of HPV DNA in healthy perilesional skin in both immunocompromised and immunocompetent patients is consistent with the findings of previous studies, suggesting that HPV DNA is widely distributed in the skin of the general population.18,19 Studies of immunocompetent patients have found HPV DNA in 67% to 74% of plucked hairs19,20 and in 69% to 80% of skin swabs.21 These findings, together with our data, suggest that the presence of HPV DNA is widespread in normal-appearing skin. In other studies, however, HPV DNA was detected more frequently in lesional versus healthy perilesional skin. A study by Forslund and colleagues22 compared the presence of HPV in NMSC, actinic keratoses, and healthy skin. They found a higher prevalence of HPV in the tumor samples than in healthy skin and concluded that the β-PV species 2 is associated with SCC. In our study we did not determine the presence of HPV in actinic keratoses and the majority of tumors analyzed were BCC. We should point out that most studies have linked HPV infection with SCC.23 Fewer studies have attempted to establish a link between HPV infection and BCC,24,25 and the majority have found no statistically significant association.
We found no significant differences between the 2 populations in the spectrum of HPV types, although the β-PV genus, particularly β-PV species 1, predominated. Similar findings have been reported in other articles,26 although many of those studies use the classical HPV taxonomic system, whereby HPV is classified as mucosal, cutaneous, or epidermodysplasia verruciformis (EV)-associated.27 According to the classification system proposed by de Villiers and coworkers4 in 2004 (which describes 16 HPV genera), the types previously classified as HPV-EV now form part of the β-PV genus (β-PV species 1 and 2). A study by Asgari and colleagues8 found β-PV 2 to be the most prevalent species, although in that study, as in ours, the type most often detected was HPV-20 (belonging to β-PV species 1).
Samples taken from sun-exposed areas showed a higher prevalence of HPV DNA in both immunocompromised and immunocompetent patients. Our findings support the theory of a strong interaction between UV light and the presence of HPV. UV radiation has also been associated with NMSC in β-PV-positive patients.28 Our analysis of potential confounding variables revealed that the only factor associated with an increased risk of HPV in immunocompromised patients was tumor location in an sun-exposed area. In immunocompetent patients a high level of occupational sun exposure, tumor location in an exposed area, and low phototype were also significantly associated with an increased risk of HPV. An important role of UV radiation has also been proposed by Forslund and coworkers,22 who found that HPV DNA levels were significantly associated with sun exposure, both in lesional and healthy skin.
Analysis of the presence of HPV in tumor samples revealed a higher prevalence of HPV DNA in SCC in situ samples than in SCC or BCC samples in both immunocompromised and immunocompetent patients. This result is consistent with other findings supporting the theory that in skin cancer HPV is more frequently detected in the earliest lesions, such as SCC in situ and keratosis with dysplasia, and would suggest that HPV primarily influences the initial stages of skin carcinogenesis rather than tumor perpetuation.29,30 Although we did not determine the presence of HPV in actinic keratoses, the presence of actinic keratoses or warts was associated with an increased risk of HPV in immunocompromised patients. However, this association was not statistically significant, probably due to the small sample size.
The data presented here highlight potentially important differences in HPV prevalence between immunocompromised and immunocompetent patients, supporting the existence of a complex interaction between HPV, exposure to UV radiation, and immunosuppression. The higher prevalence of HPV found in healthy perilesional skin suggests that HPV DNA is widely distributed in the general population. Although we found no correlation between the presence of HPV and skin cancer, it is important to note that this was a small, informative series, and that larger studies are needed to evaluate the role of HPV in skin cancer.
Ethical DisclosuresProtection of Persons and AnimalsThe authors declare that no experiments were performed on humans or animals during the course of this study.
Data ConfidentialityThe authors declare that they have followed the protocols of their place of work pertaining to the publication of patient data and that all patients included in the study were appropriately informed and provided written informed consent to participate in the study.
Right to Privacy and Informed ConsentThe authors declare that no private patient data appear in this article.
Conflict of InterestThe authors declare that they have no conflict of interest.
We thank the Department of Pathological Anatomy and Microbiology at the University Hospital Doctor Peset, Valencia, Spain, for their assistance in the preparation of this study.
Please cite this article as: Bernat-García J, Morales Suárez-Varela M, Vilata-Corell JJ, Marquina-Vila A. Detección del virus del papiloma humano en muestras de cáncer cutáneo no melanoma y piel sana perilesional en pacientes trasplantados renales y pacientes inmunocompetentes. Actas Dermosifiliogr. 2014;105:286–294.