Melanoma remains one of the most aggressive forms of cutaneous malignancies. While its diagnosis based on histologic parameters is usually straight forward in most cases, distinguishing a melanoma from a melanocytic nevus can be challenging in some instances, especially when there are overlapping clinical and histopathologic features. Occasionally, melanomas can histologically mimic other tumors and even demonstration of melanocytic origin can be challenging. Thus, several ancillary tests may be employed to arrive at the correct diagnosis. The objective of this review is to summarize these tests, including the well-established and commonly used ones such as immunohistochemistry, with specific emphasis on emerging techniques such as comparative genomic hybridization, fluorescence in situ hybridization and imaging mass spectrometry.
Los melanomas continúan siendo unas de las neoplasias cutáneas más agresivas. Si bien su diagnóstico por medio de parámetros histológicos suele ser sencillo en la mayoría de los casos, la distinción entre un melanoma y un nevo melanocítico puede suponer un reto en ocasiones, sobre todo cuando las características histopatológicas y clínicas se solapan. A veces los melanomas pueden imitar histológicamente a otros tumores, e incluso demostrar el origen melanocítico resulta complicado. Por tanto, se deben realizar varias pruebas complementarias para alcanzar el diagnóstico correcto. El objetivo de la presente revisión es resumir dichas pruebas, entre las que se incluyen algunas de uso habitual como la inmunohistoquímica, enfatizando de manera específica en las técnicas emergentes como la hibridación genómica comparativa, la hibridación in situ con fluorescencia y la espectrometría de masas.
Accurate diagnosis of melanoma is the most important prerequisite for optimal clinical management. Examination of hematoxylin and eosin (H&E) stained sections using conventional microscopy remains the gold standard for diagnosis of melanocytic lesions and as such most of the criteria used in the American Joint Committee on Cancer (AJCC)/College of American Pathologists (CAP) recommendations for reporting specimens with melanoma are based on routine examination of histologic slides.1 However, when lesions exhibit architectural and cytologic features that overlap both melanocytic nevi and melanomas, histologic examination alone may be insufficient for diagnosis. Being one of the ultimate histologic mimickers, melanoma may present in a range of histologic variants with spindle cell, desmoplastic, clear cell, balloon cell, signet ring cell, small cell, plasmacytoid and rhabdoid phenotypes.2–4 Rarely, even establishing melanocytic lineage of a tumor may become a challenging task, especially when the tumor cells are poorly differentiated and resemble multiple other tumors such as carcinoma, lymphoma, neuroendocrine carcinoma, angiosarcoma, fibrohistiocytic, and small round blue cell tumors.5 The objective of this article is to review the various ancillary tests that are available to pathologists that assist in the diagnosis of melanocytic lesions as well as their possible application to determine prognosis and therapy.
ImmunohistochemistryImmunohistochemistry (IHC) is arguably the most commonly employed ancillary test in pathology. This staining technique is used to detect the presence or absence of an antigen usually a protein.6,7 Introduced in the early 1940s, the current IHC protocols utilize specific antibodies tagged with a visible label or chromogenic agent against the target molecule.8 The presence of the antigen is revealed by serial amplification of the initial signal, thus enabling visualization of the protein and its distribution within the various cellular and extracellular components of the tissue as well as within the subcellular compartments.
Diaminobenzidine (DAB) is the most commonly used chromogen, which produces a brown precipitate. However, in heavily pigmented lesions, distinguishing the immunoreaction from melanin pigment can be challenging; therefore, other chromogens may be used including fast red or 3-amino-9-ethylcarbazole (AEC) or Texas red. An alternate approach would be to use a counterstain other than hematoxylin such as azure B or Giemsa to convert the brown pigment of melanin to green.9 Such method allows distinction between the brown DAB and the now green-colored melanin. Bleaching of melanin using potassium permanganate solution may affect the antigenicity of some epitopes and thus, affect immunodetection of certain markers.9 However, bleaching using hydrogen peroxide may be a better alternative.10 But we recommend avoiding such techniques.11
Though many of the common melanocyte specific markers are useful in the diagnosis of melanocytic lesions and primary melanomas,11,12 their expression may be lost focally or diffusely as the disease progresses, particularly in metastatic lesions. In addition, there might be acquired variable and aberrant expression of non-melanocytic markers such as cytokeratins, and neuroendocrine markers.13–15 This altered immunohistochemical profile may be accompanied by unusual tissue morphology as well. Therefore, one must exercise caution when examining a poorly differentiated neoplasm in a patient with prior history of melanoma. The following section will summarize commonly used diagnostic and prognostic markers of melanoma, not including the less commonly utilized markers such as PNL2, KBA.62, SM5-1, MC1R, NKI/C3, CSPG4 and CD146.5,16,17
Markers of melanocytic differentiationS100BSince its expression in melanoma cell lines was documented 35 years ago, S100B, also referred to as just ‘S100’, is the most analyzed marker of melanocytic differentiation.18–21 It is a member of the S100 family of calcium binding heteromeric proteins that is expressed in a variety of tissues including melanocytes.22 In addition to being expressed intracellularly, and thus, being detectable by IHC, S100 is also secreted in the serum.23 The sensitivity of S100 in melanomas is very high in formalin-fixed tissues and thus, more than 90% of all primary melanomas, including desmoplastic variants, express S100 protein.24,25 However, its expression may be lost in metastases as well as in some spindle cell/desmoplastic melanomas.26,27 Also, since the specificity of S100 is not high, it is advisable to use it in conjunction with other melanocytic markers. Regarding tissue processing, S100 protein is soluble in organic solvents such as acetone and alcohol. Therefore, frozen tissue sections and cytology preparations utilizing alcohol based fixatives are likely to yield false negative results and are not recommended for S100 IHC.
HMB45Human Melanoma Black-45 (HMB-45) is a mouse monoclonal antibody that recognizes gp100 or Pmel17, a premelanosome protein,28,29 which makes HMB45 very specific marker for melanocytic proliferations. Expression of HMB45 is directly proportional to melanin synthesizing capacity of the cells; therefore, melanocytes of blue nevi and some Spitz nevi exhibit uniform staining throughout the lesion,30 whereas the maturing dermal melanocytes of banal compound and intradermal nevi progressively lose their HMB45 expression.12,31 Most of the primary epithelioid melanomas, on the other hand are typically characterized by patchy expression of HMB45, while a majority of the desmoplastic melanomas tend to be negative for HMB45.32,33 As such, diffuse expression through the lesion or limited to the intra- and periepithelial melanocytes is usually associated with a diagnosis of nevus while patchy expression is more likely to be seen in melanomas.
MART1Melanoma antigen recognized by T cells-1 (MART1/Melan-A) was identified as one of proteins recognized by tumor-infiltrating lymphocytes.34 MART1 interacts with gp100 and plays an important role in trafficking of gp100 to premelanosomes.35 It is one of the most widely used markers of melanocytic differentiation, recognized by 2 clones of antibodies: M2-7C10 and A103.36,37 MART1 is expressed by almost all benign melanocytic lesions as well as by the majority of epithelioid melanomas.38,39 However, desmoplastic melanomas are predominantly negative for MART1.38 One of the drawbacks consistently seen with MART1 is the over-estimation of junctional melanocytes in chronically sun-damaged skin.40–42 This is due to localization of MART1 to melanosomes and the plasma membrane, in addition to the Golgi apparatus and other organelles and thus, to the dendritic processes of the melanocytes.43 Therefore, extreme caution must be employed while interpreting intraepidermal pigmented lesions studied solely with anti-MART1.42
TyrosinaseTyrosinase is a copper-containing metalloenzyme that catalyzes three reactions in the melanin synthetic pathway, including hydroxylation of tyrosine.44Tyrosinase gene encodes for peptides recognized by cytolytic T-cells in melanoma patients.45 Tyrosinase is expressed in almost all epithelioid melanocytic lesions in a finely granular pattern within the cytoplasm. Though most melanocytic nevi express tyrosinase, the intensity of staining may decrease gradually as the melanocytes descend into the deep dermis.46 It is expressed in most epithelioid melanomas, where the pattern can be patchy, but the labeling is frequently weak, focal or absent in melanomas with spindled melanocytes and in metastatic melanomas.47,48 In general, tyrosinase is comparable to HMB45 in terms of expression and usefulness in the diagnosis of melanocytic lesions.12
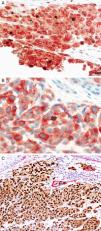
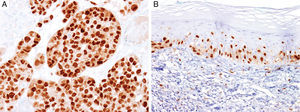
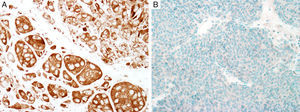
MITFMicrophthalmia-associated transcription factor (MITF) is a dimeric transcription factor that plays a seminal role in melanocyte development, proliferation, function and survival.49–51 Though expressed in several other cells such as macrophages, fibroblasts, smooth muscle cells, Schwann cells and mast cells, the nuclear localization of this protein offers a major advantage in the detection of melanocytes52. This is particularly useful when the lesion is densely pigmented. Melanocytic nevi, epithelioid melanomas as well as metastatic melanomas with epithelioid morphology express MITF, while spindled and desmoplastic melanomas are typically negative (Fig. 1A).53–55 One of principal uses of MITF is the accurate enumeration of melanocytes in intraepidermal melanocytic proliferations to distinguish between pigmented actinic keratosis from lentigo maligna (Fig. 1B).56,57
Immunohistochemical study for microphthalmia-associated transcription factor (MITF). (A) Invasive melanoma diffusely positive nuclear labeling in melanocytic lesions with epithelioid melanocytes (magnification 200×). (B) Melanoma in situ, with contiguous proliferation and suprabasal spread of atypical melanocytes (magnification 200×).
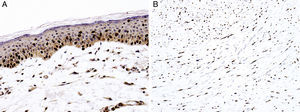
Sex determinant region Y (SRY) related HMG-box gene 10 (SOX10), is a transcription factor that binds to and transactivates MITF gene promoter and regulates the expression of tyrosinase-related protein-1 among others and plays a critical role in melanocyte growth and function.58,59 SOX10 is a nuclear marker that can be expressed in Schwann cells, eccrine epithelium, myoepithelial cells and glial cells.60,61 Similar to MITF, SOX10 is a useful marker for the distinction of pigmented actinic keratosis from melanoma in situ (Fig. 2A).57. It is also more sensitive than MITF in that almost all spindle cell melanomas and most desmoplastic melanomas express SOX10 (Fig. 2B)60,62,63.
p75NGFRNerve growth factor receptor (molecular weight: 75kDa), also known as neurotropin receptor and CD271 belongs to the tumor necrosis factor receptor superfamily and is widely expressed in the skin. Transit amplifying keratinocytes of basal layer, follicular outer root sheaths, myoepithelial cells, fibroblasts and nerve fibers express NGFR.64,65 It is not expressed in epithelioid type-A melanocytes; but is induced during neurotization and as such is expressed in melanocytes with spindled morphology (type-C). NGFR has a higher sensitivity for detection of spindle cell and desmoplastic melanomas and has been touted to be a better marker than S100 for spindle cell melanomas.27 In addition, other spindle cells tumors of sun-damaged skin are frequently negative for NGFR.66,67 However, caution must be exercised while using NGFR as a sole marker of desmoplastic or spindle cell melanoma, since it can be expressed in dermatofibrosarcoma protuberans and scars.68,69
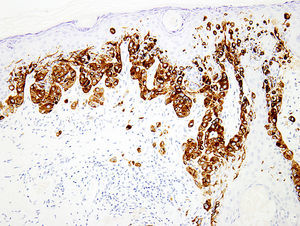
Melanocytic marker cocktailsSometimes, a mixture of 2 or more monoclonal antibodies against melanocytic markers is used in immunohistochemistry.70,71 Though many of these cocktails are generated and validated in individual laboratories, some are also commercially available.16 The main advantage of this is the increased sensitivity for detection of melanocytes. For instance, combination of HMB45 and anti-tyrosinase can be used to determine the number and growth pattern of intraepidermal melanocytes and for detecting the presence of a dermal component as well (Fig. 3). The main application of such cocktails is in establishing a melanocytic origin in a given neoplasm and in the detection of microscopic metastases in sentinel lymph nodes.
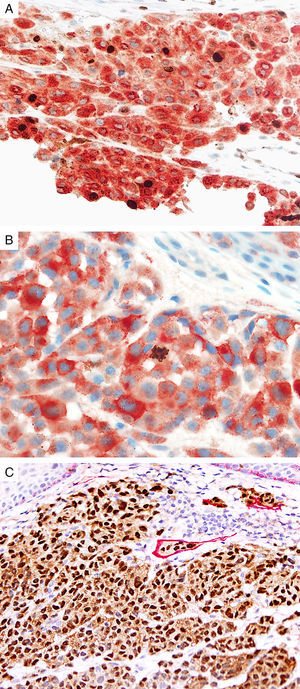
Prognostic markers of melanomaKi67Marker of proliferation Ki67 is a nuclear non-histone protein that is expressed in actively proliferating cells (not in G0 phase).72 The proliferative activity, as indicated by the percentage of lesional cells expressing Ki67 could be extrapolated to determine the biologic behavior of the melanocytic lesion. In general, fewer than 5% (usually 1%) of melanocytes comprising most benign lesions expressed Ki67,12,73,74 with a decreasing gradient toward the deeper aspect, reflecting maturation of melanocytes with progressive dermal descent.75 In melanomas, the Ki67 proliferation index is higher, ranging from 5% to 50%, with mitotic figurees distributed throughout the lesion.75 In some cases, foci of increased proliferation may be seen. Increased Ki67 proliferation rate has been correlated with recurrence and metastasis, particular in thick melanomas.76–78 To help distinguish between proliferating melanocytes and other cells at our institution, the growth fraction is determined by adding a cytoplasmic melanocytic marker such as MART1/Melan-A/tyrosinase to anti-Ki67 (Fig. 4A). This combination is particularly useful in lesions with a brisk inflammatory infiltrate. In addition, Ki67 positivity of intraepidermal melanocytes is seen more commonly in in situ melanomas, compared to nevi.79
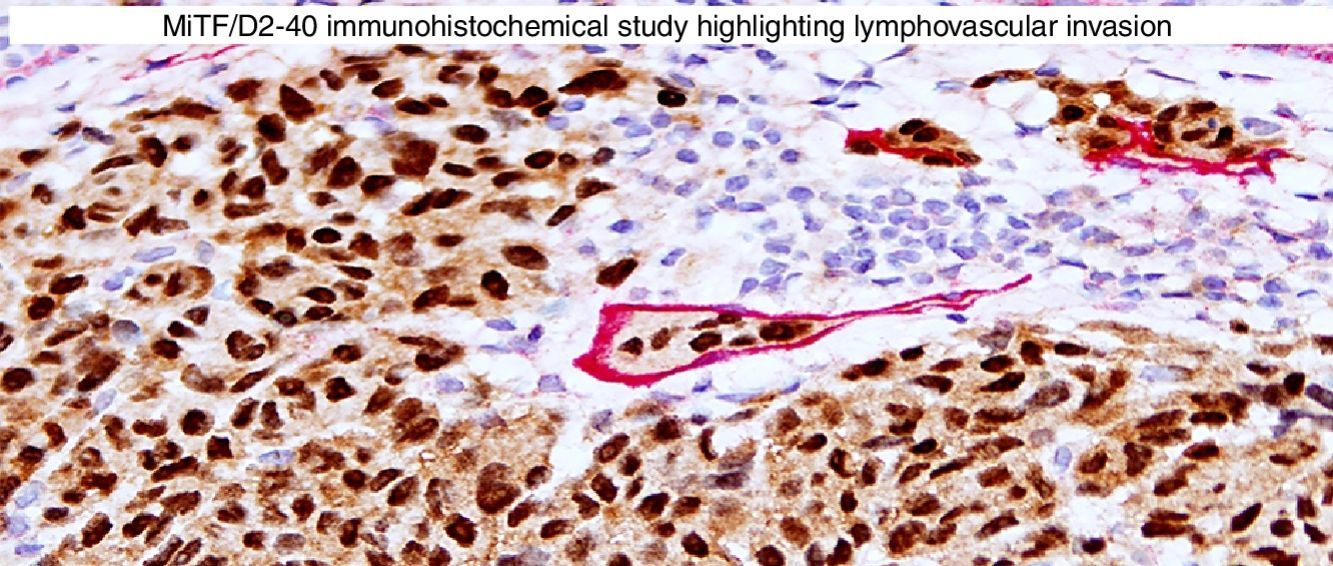
Immunohistochemical studies for prognostic factors. (A) Mart-1/Ki67 cocktail highlights the melanoma cells (red, cytoplasmic) with increased nuclear positivity for Ki67 (brown, nuclear) (magnification 200×). (B) Mart-1/PHH3 cocktail highlights a mitotic figure (brown) in a melanoma (red, cytoplasmic) (magnification 200×). (C) MITF/D2-40 reveals the presence of lymphovascular (red) space invasion by melanoma cells (brown, nuclear) (magnification × 100).
Identification of mitotic figures is an important staging parameter in thin melanomas (Breslow thickness≤1.0mm).80 Phosphorylation of histone H3 begins in the late G2 phase of cell cycle. Phosphohistone H3 (pHH3) can be detected throughout the entire M-phase and thus, is a marker of cells undergoing mitosis.81–83 pHH3 expression in dermal melanocytes has been correlated with outcomes in melanoma.84–87 While manually identifiable mitotic figures do not include cells in prophase, pHH3 detects these cells as well. The AJCC-TNM (2009) staging system is based on mitotic figures detected on routine, H&E-stained slides, and thus immunedetection of pHH3 should not be used in routine examination of primary melanomas. Anti-pHH3 antibody may also be combined with a cytoplasmic melanocytic marker such as MART1 to enhance detection of actively dividing melanocytes (Fig. 4B).84,88 At our institution we use such a combination of anti-MART1 and anti-pHH3 in cases where we identify cells undergoing mitosis but we are not certain if they correspond to a melanocyte or to other dermal cells (e.g., endothelial cells, lymphocytes, macrophages).
D2-40D2-40 is a monoclonal antibody that recognizes podoplanin, a transmembrane mucoprotein that is selectively expressed in lymphatic endothelium.89,90 Presence of lymphovascular space invasion (LVI) by melanoma has been correlated with metastases to sentinel and regional lymph nodes, and decreased disease-free and overall survival.91–93 Detection of LVI by examination of H&E stained sections alone may be difficult since filling up of small vessels with tumor cells may lead to flattening of endothelium, while retraction artifacts and dyshesive properties of the tumor cells may mimic a vascular space.94 Therefore, the use of lymphatic and vascular endothelial markers has become a common practice, particularly in thick melanomas. Dual immunostaining using S100/D2-40 cocktail did not correlate with sentinel lymph node status.95 However, a recent study from our group revealed that detection rates of LVI were increased by using MITF/D2-40 cocktail with positive correlation with sentinel lymph node metastasis (Fig. 4C).96
BRAFV600ESomatic missense mutations in BRAF, a cytoplasmic serine/threonine kinase are common in cutaneous melanomas (50–60%).97 A single nucleotide mutation 1799 T>A constitutes more than 90% of these mutations, resulting in substitution of glutamic acid for valine (BRAF V600E). With the advent of BRAF inhibitors, detection of this mutation has acquired a paramount importance in management of patients with metastatic melanoma.98,99 Though the gold standard is molecular testing, immunohistochemical studies using anti-BRAFV600E antibody (clone VE1) have demonstrated high concordance with the results of molecular analysis.100–103 The staining is usually diffuse and cytoplasmic (Fig. 5); but, focal staining is also observed in several cases,83 which could be attributed in part to technical issues.104 Nevertheless, other studies have demonstrated that the heterogeneous BRAF V600E staining could indeed be a reflection of the polyclonal nature of melanoma.105 Caution must be exercised when analyzing the slides, especially in densely pigmented lesions, where the cytoplasmic melanin pigment may mimic positive staining. In such cases, counterstaining with Giemsa is warranted in order to avoid a false positive reading.106 Recent reports have demonstrated that the immunostain might be too sensitive in some cases,107 so more studies may be needed to determine the clinical application of immunohistochemical detection of BRAF V600E.
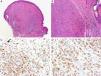
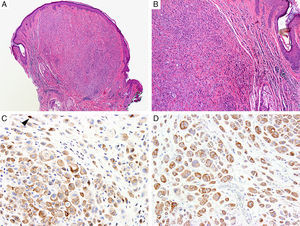
BAP1Breast cancer type 1 susceptibility protein (BRCA1) associated protein-1 (BAP1), is a ubiquitin carboxy-terminal hydrolase that functions as a tumor suppressor.108,109 Individuals with heterozygous germline BAP1 mutations are at high-risk for developing a variety of tumors, including asbestos-associated mesothelioma, lung adenocarcinoma, meningioma, and renal cell carcinoma.110–112 These patients may also develop a gamut of melanocytic proliferations ranging from epithelioid nevi to spitzoid melanocytic proliferations (the so-called ‘BAPomas’) to melanomas (cutaneous and uveal).113–115 These tumors are characterized by loss of nuclear BAP1 staining, usually accompanied by of BRAF V600E expression (Fig. 6).116–119 Though BAP1 has traditionally been considered a tumor suppressor, recent studies have reported that BAP1 might play a role in survival of tumor cells.120
Immunohistochemical studies for BAP1 and BRAF in a ‘BAPoma’. (A) Hematoxylin and eosin stained section of a dome-shaped BAPoma with dense proliferation of epithelioid melanocytes within the dermis (magnification 40×). (B) Typical nevus cells are also present at the peripheral of epithelioid proliferation (right side, magnification 100×). (C) Diffuse loss of nuclear BAP1 in most of the melanocytes; arrowhead indicates single positive cell. Of note, the background cytoplasmic reaction should not be interpreted as positive (magnification 200×). (D) These cells are diffusely positive for BRAF V600E protein (magnification × 200).
While most melanocytic lesions can be characterized by microscopic examination of H&E-stained sections and immunostains, some melanocytic proliferations presenting with ambiguous features can be difficult to diagnose accurately. In such cases, molecular studies may provide additional diagnostic information. Though cutaneous melanomas represent a divergent group of diseases, they usually have recurrent, non-random chromosomal abnormalities,121,122 and detection of these aberrations may aid in the diagnostic process.123 The following section will summarize the basic principles and practical application of the various molecular tests for the diagnosis of melanocytic lesions.
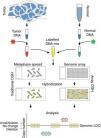
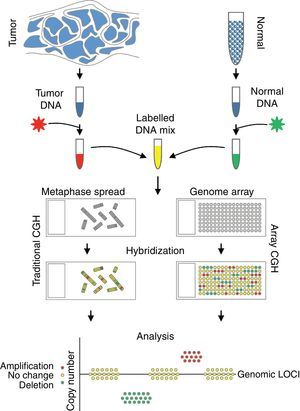
Comparative genomic hybridizationComparative genomic hybridization (CGH) is a cytogenetic assay that can detect losses and gains of genomic material and map them to specific chromosomes.124 CGH utilizes differential labeling of genomic DNA extracted from fresh or formalin-fixed tumor and normal tissue (usually peripheral blood leukocytes) with fluorochromes, followed by co-hybridization of equal amounts of each fraction on to metaphase spreads of chromosomes derived from a peripheral blood lymphocytes of a healthy donor (traditional CGH). DNA copy number changes can be determined by comparison of relative signal intensities of the different fluorochromes (Fig. 7). The main advantage of CGH is the ability to analyze the entire genome simultaneously and objectively, without specific probes. However, the utility of traditional CGH is restricted due to limited resolution of 3–10Mb.125 This can be overcome by utilizing an array of DNA targets covering the entire genome for hybridization (array CGH).126,127
The potential of CGH in the classification of melanocytic lesions began with the work of Bastian et al., who analyzed 32 primary melanomas.128 They identified loss of chromosome 9, particularly involving the short arm in majority of the melanomas. In addition, losses of parts of chromosomes 9 and 10 occurred in early lesions, whereas gains of chromosome 7 were late events in melanoma progression. A subsequent study from the same group revealed that chromosomal alterations could be identified in both melanomas and melanocytic nevi, albeit in a lower frequency and particularly in Spitz nevi. While cytogenetic alterations seen in melanomas involved portions of chromosomes, those in nevi typically involved the whole chromosomes or entire arms of the chromosomes.129,130
Metastatic melanomas have been shown to have a distinct set of chromosomal abnormalities, compared to primary melanomas.131 Some studies have revealed that loci of tumor suppressors are frequently deleted while those containing proto-oncogenes may be amplified.132–134 Among the amplified genes are BRAF and NRAS,134 somatic mutations of which have been implicated in early melanomagenesis. Melanomas of various histologic subtypes may be grouped together based on their cytogenetic abnormalities.135 For example, melanomas that arise in sun-exposed regions may exhibit NRAS and KIT mutations while most superficial spreading melanomas show BRAF mutations.
In addition to aiding in the diagnosis, CGH can also predict the biologic behavior of melanoma to some extent.123 Gains of 6p were seen exclusively in thicker melanomas, which along with gains at 1q were associated with a poor outcome.136 The total number of genomic aberrations in a tumor has been correlated with outcome: the higher the number of chromosomal alterations, the worse the prognosis. Some studies have demonstrated that the frequency of homozygous deletions might be a better predictor of metastasis to sentinel lymph nodes and distant sites.137
Thus, CGH may be a useful tool that can aid in distinguishing benign melanocytic proliferations from melanomas and, to some extent in predicting the outcome. However, the utility of CGH is limited by requiring a relatively high and “pure” number of tumor cells in the sample. Lesions with admixed tumor infiltrating lymphocytes, other inflammatory cells, and prominent intervening stroma are less frequently successfully studied by CGH. Also, when only a fraction of the cells (<30%) contain the cytogenetic abnormalities, they may evade detection by CGH, thus yielding a false negative result.130 In addition, the presence of balanced translocations cannot be detected by CGH.
Fluorescence in situ hybridizationFluorescence in situ hybridization (FISH) is a cytogenetic assay used to determine the copy number of specific genomic regions.138 Its application in the diagnosis of melanocytic lesions was realized only recently, after high-throughput use of CGH proved to be challenging for routine clinical use. In this assay, paraffin sections representing the tissue of interest are hybridized with a mixture of differentially labeled fluorescent probes targeting specific cytogenetic loci. The corresponding signals are enumerated per cell as the percentage of lesional cells containing altered number of signals. The advantages of FISH over CGH include the ability to examine the tumor cells specifically for the number of signals and thus, the feasibility for application in lesions with smaller tumor volume. However, since examination of immunofluorescence slides provides less defined morphologic detail than routine sections, dense lymphohistiocytic infiltrate and nevoid morphology of the tumor cells may result in difficult quantification of the FISH signals.
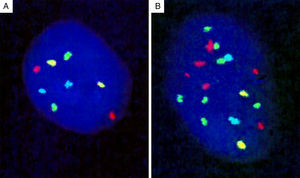
Genomic loci tested by FISH represent regions with the most frequent chromosomal aberrations, and 13 such hotspots were initially identified by analysis of the CGH assays.139 Of these, combination of four different probes produced the most distinct separation of melanomas from other melanocytic lesions. The initial 4-probe FISH assay utilized probes that target the following loci on chromosomes 6 and 11: 6p25 (RREB1, red), 6q23 (MYB, gold), 11q13 (CCND1, green), and centromere 6 (Cep6, aqua) (Fig. 8). A lesion was considered to be melanoma if at least one of the following chromosomal abnormalities was present: (i) 3 or more green dots per nucleus (11q13, CCND1) in at least 39% of tumor cells, (ii) 3 or more red dots (6p25, RREB1) in at least 30% of tumor nuclei, (iii) number of RREB1 signals (red dots) greater than the number of centromere 6 signals (aqua dots) in more than 55% of tumor nuclei and (iv) at least 41% of tumor cells with fewer number of MYB signals (gold dots), compared to centromere 6 signals (aqua dots). Using these criteria, nevi could be distinguished from melanomas with high sensitivity (90%) and specificity (95%). Of these, gain of 6p25 was found to have the highest sensitivity for this distinction.140,141
Four-probe FISH assay. (A) Normal cell with two signals each of 6p25 (RREB1, red), 6q23 (MYB, gold), 11q13 (CCND1, green), and centromere 6 (Cep6, aqua). (B) Melanoma cell with an abnormal FISH result with multiple RREB1 (6 red dots) and CCND1 signals (7 green dots), indicating gains at 6p25 and 11q13, respectively.
The applicability of FISH with the 4-probe method has been examined in various settings including pigmented melanocytic lesions with blue nevus like morphology142,143; lesions with desmoplastic response144; lesions with nevoid melanocytes145,146; conjunctival melanocytic lesions147 and even intranodal melanocytic deposits148, as well as in distinguishing melanoma cells from associated nevocytes146. In addition, it is unknown how useful FISH is in ambiguous melanocytic lesions since most studies were done in histologically obvious nevi or melanoma and the presence of cytogenetic alterations did not always correlate with the clinical outcome.139,149–152 Furthermore, the presence of tetraploidy and polyploidy (common in Spitz and other melanocytic nevi) may yield false positive results.151,153–155
Additional genomic loci initially identified by CGH were examined to address the lack of sensitivity of conventional 4-probe FISH assay with respect ambiguous melanocytic lesions and in identifying polyploidy.156 Based on this, the current FISH panel includes probes against 6p25 (RREB1, red), 9p21 (CDKN2A, gold), 11q13 (CCND1, green), 8q24 (MYC, aqua) and centromere 9 (Cep9, green). The presence of at least 30% of tumor cells with gains of RREB1 (red), CCND1 (green) or MYC (aqua) with or without homozygous deletion of CDKN2A (gold) is considered to be a positive result. Use of this probe set has increased the sensitivity from 75% to 94% for the diagnosis of melanoma, in addition to eliminating most false positive results due to tetraploidy.157 Though this is a significant improvement from the initial panel, FISH assay should be used only to supplement the findings of a thorough histopathologic examination and the results from a FISH study should not influence the management of the patient.152
Imaging mass spectrometryMass spectrometry (MS) is a technique that utilizes molecule specific mass to charge (m/z) ratios to identify and quantify substances from various sources.141 MS involves generation of gas phase ions from the substance of interest, and then subject them to progressive fragmentation. The ions are separated based on their mass and charge and their amplitude of detection as ‘peaks’ would be directly proportional to their relative concentration in the parent substance. Each molecule has a specific m/z ratio, which will allow identification of the molecular composition of the parent substance.
Imaging mass spectrometry (IMS) is a process where the MS analyses are performed directly on tissue sections. Therefore, the analysis can be restricted to specific regions of the sections, and thus, target only cells of interest with minimal contamination from the adjacent tissues.103,158 In contrast to routine immunohistochemical studies and other ancillary tests used in pathology, using IMS, the distribution and abundance of all proteins and peptides can be analyzed simultaneously. In situ imaging of proteins provides an unbiased view of molecular composition of tissues and has been used mostly in the research setting so far.159
In an attempt to facilitate accurate diagnosis of spitzoid lesions, Lazova and colleagues analyzed 114 cases,160 with 51 as the training set, and a validation set of 63 cases. Two serial sections of formalin fixed paraffin tissues were cut on to a glass side for H&E staining and on to a conductive glass slide for IMS. After marking areas of interest on the H&E slide, including dense areas of tumor devoid of other cells such as stroma or lymphocytes, the corresponding areas on the conductive slide were analyzed by IMS. Of the multiple models studied by the researchers, 5 peaks corresponding to tumor and 12 peaks corresponding to tumor microenvironment were found to provide the most discrimination between Spitz nevi (SN) and spitzoid melanoma (SMM). Tumor-associated peptides could distinguish the spitzoid lesion with a sensitivity of 97% and specificity of 90%, whereas the sensitivity and specificities of tumor microenvironment-associated peptides were 64% and 90%, respectively. Of the five differentially expressed tumor-associated peptides, the authors were able to identify actin and vimentin. Though IMS is at an early stage, it has shown promise in distinguishing between melanoma and congenital nevi.161
ConclusionMelanoma continues to be a deadly disease and accurate diagnosis is critical for appropriate treatment of the patient. The various ancillary tests mentioned in this review serve as supplements to thorough histologic examination. In addition, they can also provide valid prognostic information and identify patients that may benefit from targeted therapy.
Conflict of interestThe authors declare no conflict of interest.