The worldwide explosion of interest in artificial intelligence (AI) has created a before-and-after moment in our lives by generating great improvements in such sectors as the automotive and food production industries. AI has even been called the fourth industrial revolution. Machine learning through AI is helping to improve professional processes and promises to transform the health care sector as we know it in various ways: 1) through applications able to promote health in the general population by providing high-quality information and offering advice for different segments of the population based on prediction models; 2) by developing prediction models based on anonymized clinical data, for preventive purposes in primary care; 3) by analyzing images to provide additional decision-making support for health care providers, for improving specialist care at the secondary level; and 4) through robotics applied to processes that promote health and well-being. However, the medical profession harbors doubts about whether this revolution is a threat or an opportunity owing to a lack of understanding of AI technology and the methods used to validate its applications. This article outlines basic aspects of AI as it is applied in dermatology and reviews the main advances achieved in the last 5 years.
La irrupción de la inteligencia artificial (IA) a nivel mundial ha supuesto un antes y un después en nuestras vidas, generando grandes mejoras en diferentes sectores, como el de la automoción y el agroalimentario, entre otros, lo que ha llevado a denominarla la cuarta revolución industrial. La AI, capaz de aprender de forma automatizada y de ayudar al profesional a mejorar sus procesos, promete cambiar el ámbito sanitario tal y como lo conocemos mediante: 1) aplicaciones capaces de generar salud en la población general a partir del uso de información de calidad y de segmentación de consejos basados en modelos de predicción; 2) modelos capaces de generar algoritmos de predicción a partir de datos anonimizados procedentes de información clínica, a fin de mejorar la prevención primaria; 3) sistemas de análisis de imagen capaces de dar a los profesionales de la salud un soporte extra en la toma de decisiones, mejorando la prevención secundaria; y 4) aplicación de robótica combinada en la mejora de procesos ligados al ámbito de salud y bienestar.
Sin embargo, la falta de conocimiento tanto en este tipo de tecnología, como en los términos y la metodología de validación de la misma, hace que la clase médica dude en si esta revolución supone una amenaza o una oportunidad para la profesión. En el presente artículo de revisión pretendemos introducir una serie de aspectos básicos de la IA aplicada a la dermatología, así como los principales avances sucedidos en este campo en los últimos 5 años.
As Marie Curie said, “Nothing in life is to be feared, it is only to be understood. Now is the time to understand more, so that we may fear less.” Often, when we are faced with an unknown quantity, this can take on a mysterious dimension due to a lack of understanding.
In this article, we aim to introduce a series of basic concepts about artificial intelligence (AI) applied to dermatology, and we will present a range of examples of how AI can be applied in our specialty.
Our aim is to encourage all dermatologists to deepen their understanding of this new subspecialty. By learning about AI in dermatology, we will be better placed to lead its development, which currently is in hands of engineers who minimize the importance of correct medical diagnosis or whose focus is inappropriate, thereby limiting the true value of this innovation.
The present article was written by physicians specialized in medical-surgical dermatology and venereology who have direct and indirect experience in this new field of health. We will draw on this experience to highlight the most relevant aspects of IA and thus illustrate what impact AI might have on the future of dermatology.
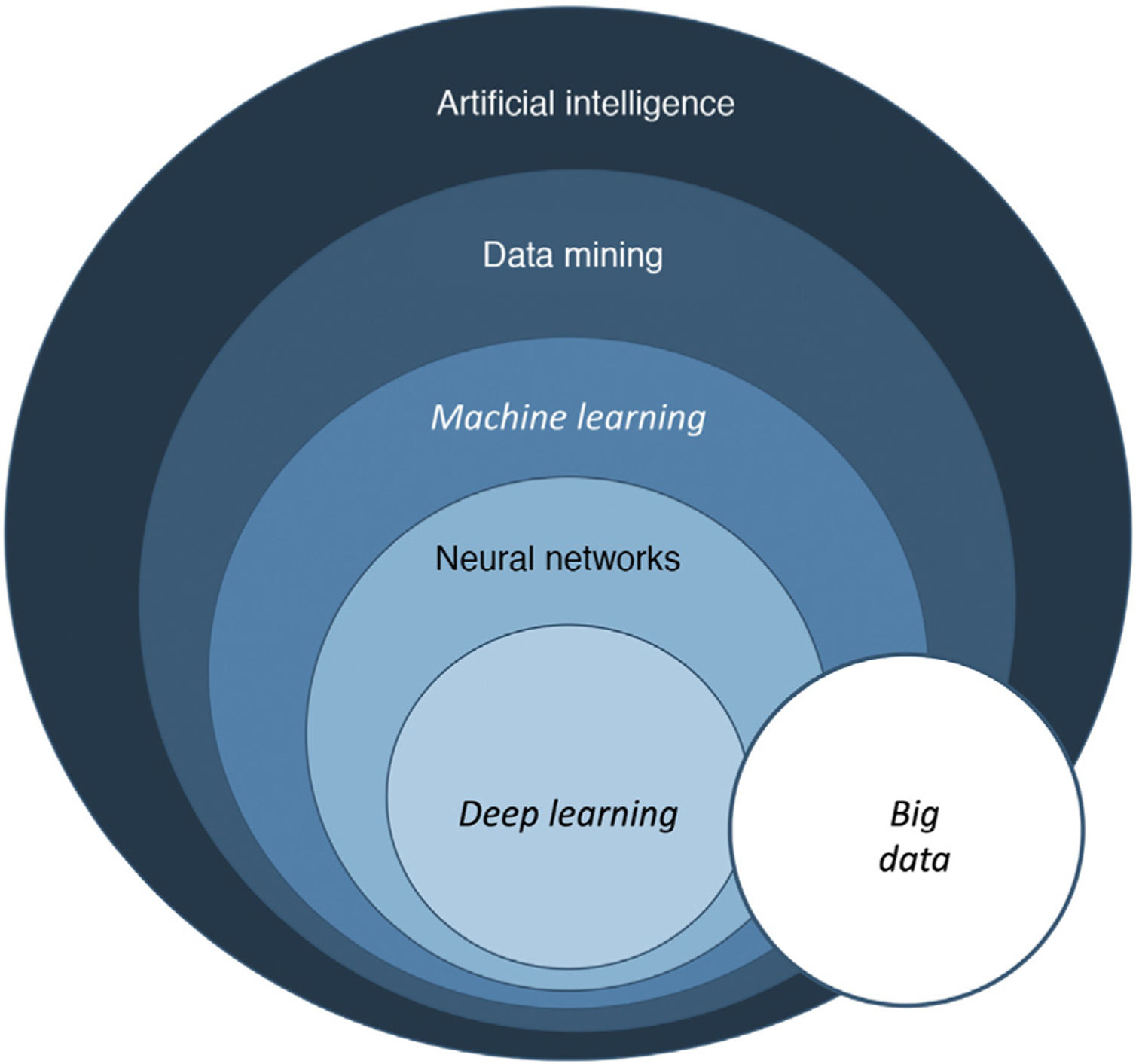
Basic notions of artificial intelligenceMedical presentations, podcasts, and interviews about AI will often use terms such as machine learning and neural networks.
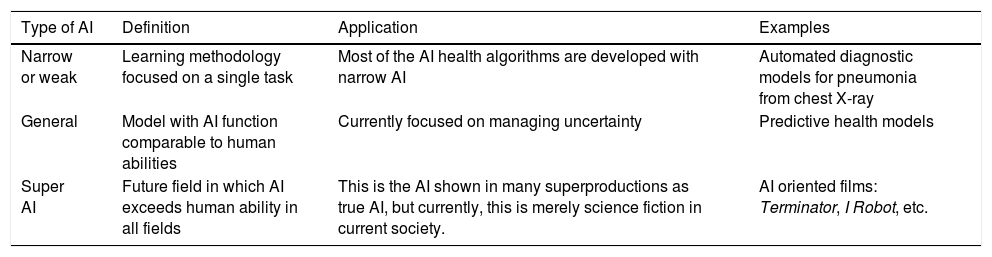
In its essence, AI is a technology developed to imitate cognitive functions of human beings1. There are 3 basic types of AI, as shown in Table 1.
Subtypes of artificial intelligence (IA).
| Type of AI | Definition | Application | Examples |
|---|---|---|---|
| Narrow or weak | Learning methodology focused on a single task | Most of the AI health algorithms are developed with narrow AI | Automated diagnostic models for pneumonia from chest X-ray |
| General | Model with AI function comparable to human abilities | Currently focused on managing uncertainty | Predictive health models |
| Super AI | Future field in which AI exceeds human ability in all fields | This is the AI shown in many superproductions as true AI, but currently, this is merely science fiction in current society. | AI oriented films: Terminator, I Robot, etc. |
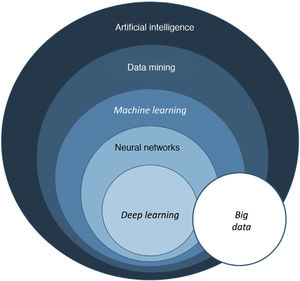
The term big data refers to data or combinations of datasets whose size (volume), complexity (variability), and rate of growth hinder acquisition, management, processing, or analysis by conventional means such as personal computers, relational databases, and conventional statistics (for example, SPSS® software)1. This type of analysis enables enormous quantities of data to be managed and generation of predictive models which are useful in clinical practice. An example would be predicting the profile of a patient who may be more susceptible to developing an infectious process in the surgical area with a view to taking preventive measures.
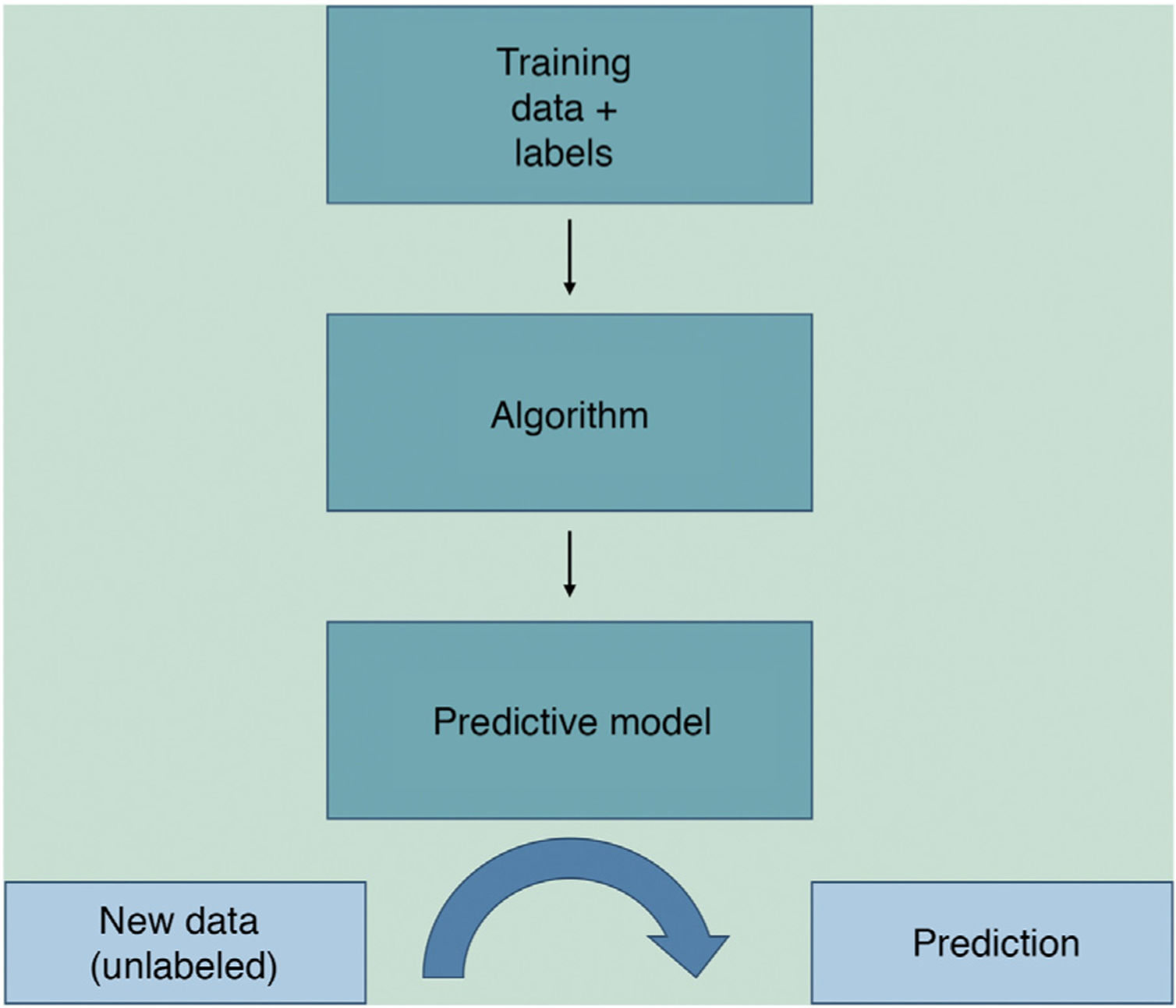
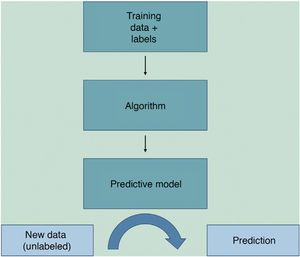
Machine learning (ML) is a type of AI that enables computers to learn from data, without being explicitly programmed (Fig. 1). This technology, which lies at the heart of AI and management of big data, has aroused great interest in recent years. In this article, we will focus on ML, as this is the most widely used modality in dermatology, along with deep learning2.
Machine learning and deep learningOne of the branches of AI with greatest potential for application in medicine is ML, which consists of independent analysis of data thanks to the generation of algorithms for decision making2–4. This technology enables the identification of patterns to classify and make different predictions.
ML works by feeding an algorithm with input data comprising past observations and constructs a model for predicting and classifying new observations unknown to the algorithm by imitating a human cognitive process (Fig. 2).
Among the most popular ML models in the health sector are convolutional neural networks (CNNs), which imitate brain neuron function. CNNs are formed of variable layers interconnected by weights that the algorithm calculates and adjusts through iterations (repetitions) in a process known as gradient descent.
In dermatology, we usually use multiple variables, and so multiple neurons are needed to weave a network of connections that generate useful information. When we talk of a system that bases its information on more than one layer of information, we refer to deep learning (DL). Such networks generate excellent results provided that 2 fundamental requirements are met: data (both images and clinical information) are available in sufficient quantity and quality (big data) (Fig. 3).
Learning models for artificial intelligenceAI (ML, DL) can essentially be trained in 3 modes: supervised, unsupervised, and by reinforcement.
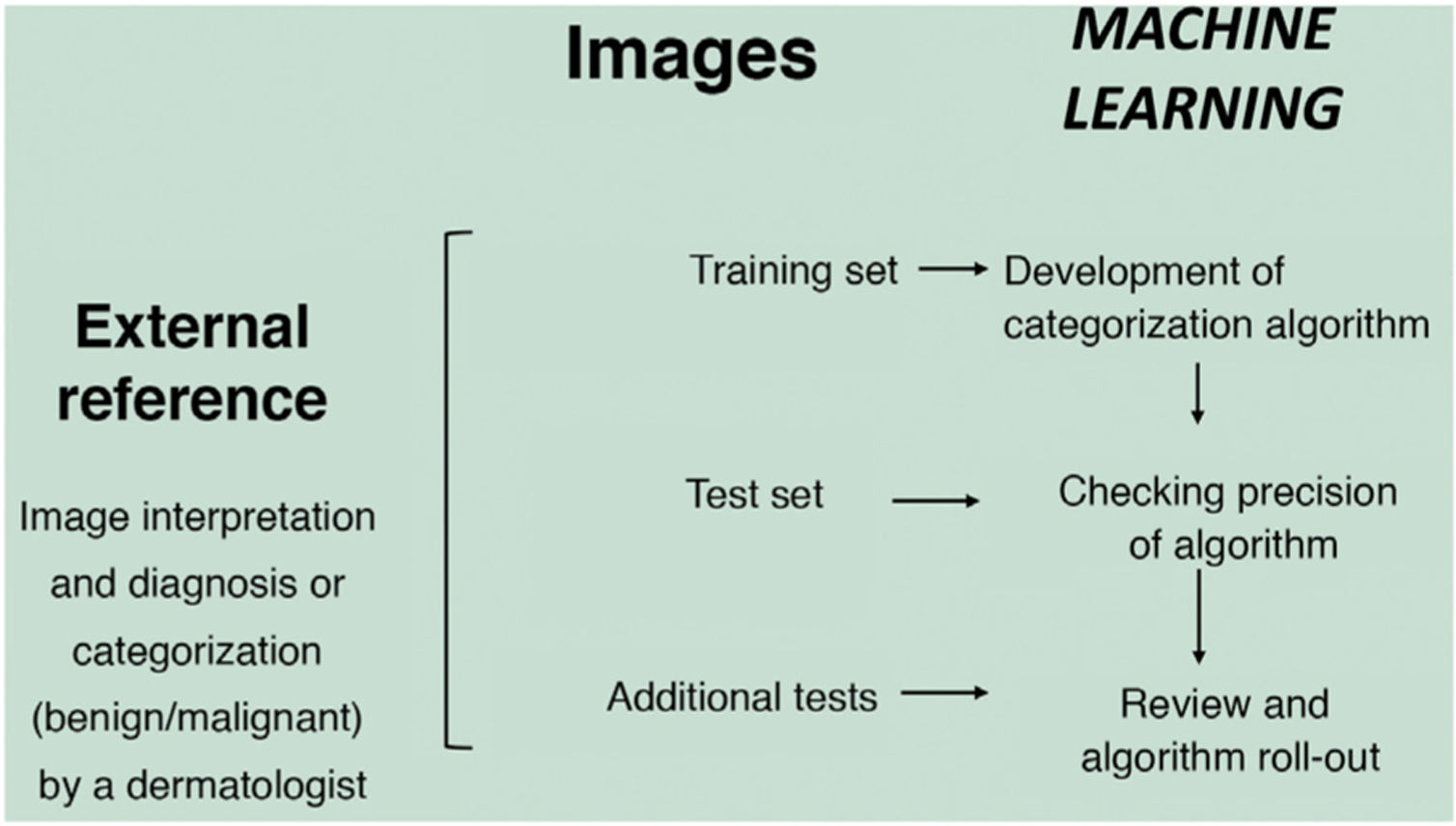
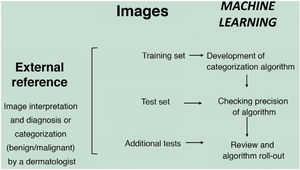
In the supervised model of learning, the AI algorithm is trained with a set of examples labelled by an expert in which the output results are known. This is the model that we usually find in dermatological applications, whose training content are images labelled with their diagnosis or selected training parameter5,6. By way of example of this supervised learning model in the field of dermatopathology, in the first phase, the machine is taught to recognize features and specific patterns, and regions of interest, validated in this case by a dermatopathologist (for example, features and patterns associated with benign melanocytic lesions and malignant ones [Fig. 4]). These features and basal patterns constitute what is known as the ground truth. In the second phase, thanks to DL algorithms, the machine is able to recognize patterns and regions of interest in lesions not previously analyzed, and diagnose a lesion as a benign or malignant lesion based on these algorithms4,5.
The unsupervised model is based on unlabeled data in which the output structure and results are not known a priori. Basically, such algorithms search for grouping patterns (clusters) for subjects that, in view of their characteristics in the different variables that define them, show similarities7. There are few applications of this type of AI in dermatology, given that in most cases algorithms are applied to images. This type of model is usually used in situations in which predictions are required or in the case of needing rapid classifications for data mining8. An example of unsupervised learning would be the generation of classifiers for ‘melanoma versus nonmelanoma’ by training a model that groups/classifies images through patterns detected by the algorithm itself8.
Artificial intelligence applied to inflammatory diseasesDL algorithms have been used to classify different inflammatory skin diseases. We will present some of the most relevant ones below.
PsoriasisAI-based algorithms can be useful both for making a more precise clinical assessment and for supporting the development of personalized therapeutic protocols and predictions of results9.
The first IA program designed for psoriasis was that created by Guo et al.10 in 2014. Their objective was to predict the development of psoriasis by microarray gene expression profiling of 2 sets of data. The binary classification achieved an overall precision of 99.81%.
Several attempts have been made to develop an AI program that could help to assess the severity of poriasis11–13. In the most recent models, the authors used 3 different standard classifiers—support vector machines, decision trees, and artificial neural networks—to stratify risk and assess 3 principal attributes: color, texture, and higher order spectra. The classifier was trained with 670 psoriasis images. The classifiers segmented the lesions and classified them as healthy or diseased. The model achieved a precision of 99.84%, a sensitivity of 99.76%, and a specificity of 99.99%. Given that the sample size was small, the result could be affected by overfitting.
Other studies aimed to develop a system to stratify psoriasis severity using the Psoriasis Area and Severity Index (PASI), assessing the affected area14,15, scaling16, induration and color17, and isolated erythema18.
New studies have managed to improve the automatic detection of psoriasis lesions through use of cluster-based segmentation along with swarm intelligence techniques19.
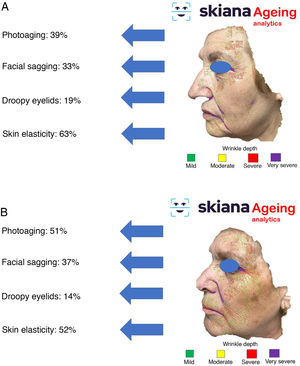
Recently, and following one of the previous models, the results were presented for the IMAPSORS project, included within the SKIANA project®. Development was cofinanced by the European Commission. The aim was to generate quantifiable information on the skin of patients with psoriasis using personal mobile devices. The results obtained in the capacity for detecting psoriasis and in automated definition of disease severity in the form of PASI and body surface area, both quantitatively and qualitatively, open the door to an improvement in the definition of severity and early detection of moderate and severe forms of the disease (Fig. 5A and B)20.
AI programs have also been developed to assess and optimize treatment of psoriasis, specifically, for determining long-term response to biologic treatment. The first of these studies was based on 2 ML models that studied gene expression in skin biopsies21. These models were able to predict PASI75 response after 12 weeks of treatment by assessing the early (2–4 weeks) expression profiling of treatment. Another study undertook multiomic analysis in patients receiving etanercept, and found indicators of response to treatment in gene expression and pathways associated with tumor necrosis factor and major histocompatibility complex signalling22.
Although studies of gene expression profiling in biopsy samples are promising, there is interest in methods that avoid the invasive nature of this technique. Thus, a predictive model has been developed using biochemical markers in blood23. However, simpler PASI-based models were better predictors and so further study is necessary.
Another recent study used ML to predict response of patients with psoriasis to biologic treatment using basic health parameters such as age of onset of psoriasis and weight of the patient24.
AI models can also be used to discover potential off-label treatments for psoriasis and other inflammatory conditions through models that use text from scientific articles and disease classifications to identify potential drugs25.
AI programs have also been used to study comorbidities in psoriasis. One of these programs identified predictors of coronary artery disease in patients with psoriasis; these predictors included obesity, dyslipidemia, and inflammation26.
Another comorbidity that affects approximately 25% of patients with psoriasis is psoriatic arthritis. To date, there is no effective method for predicting the development of this condition. An AI program has been developed based on genotypes of patients with psoriasis and psoriatic arthritis, differentiating between the 2 diseases based on 200 genetic markers, with an area under curve (AUC) of 0.82. This was the first study to show robust prediction of the development of psoriatic arthritis using only genetic information27.
Atopic dermatitisThe use of AI in atopic dermatitis can be of help both in the diagnosis and personalized treatment of the condition as well as in predicting treatment outcomes. It could also be useful for helping to standardize and reduce the time taken to assess patients.
Investigation of AI in the field of atopic dermatitis is, however, still in its infancy.
An ML algorithm has been designed to identify atopic dermatitis based on electronic health records28. For this, a natural language processor has been developed to incorporate both structured and unstructured data. Using 562 electronic health records, the designers achieved a positive predictive value of 84% and a sensitivity of 75%.
Other groups developed an artificial neural network to distinguish skin with atopic dermatitis involvement from healthy skin using imaging information29. However, the number of samples used was low, given that this was an exploratory study.
OnychomycosisA recent study compared a group of dermatologists with CNNs for the detection of onychomycosis30. A total of 49,567 images were used to train the classifier to distinguish between nails with onychomycosis and normal nails. In the validation, the classifier achieved a sensitivity of 82.7%–96.7% and a specificity of 69.3%–96.7%. The area under the receiver operator characteristics (ROC) curve was 0.82–0.98. In this study, the capacity of the classifier to differentiate healthy nails from those with onychomycosis was statistically superior to that of dermatologists.
Rosacea and acneClinical evaluation of patients with rosacea, as with other inflammatory diseases, often shows high intra- and interobserver variability. In this context, Binol et al.31 developed an AI program based on CNNs to perform a quantitative and reproducible assessment of lesions, after training with more than a million images. These authors also defined what they called regions of anatomic interest, that is, facial areas highly susceptible to rosacea, thereby significantly reducing false positives in identification of rosacea lesions.
Similarly, digital processing of images has been used for automatic detection of acne lesions, with the aim of performing a more precise and reproducible evaluation. Min et al.32 developed a system that automatically counted 5 types of lesions (papules, nodules, pustules, closed comedones, and open comedones) and compared the results with the manual count performed by a dermatologist. The lesions assessed in 25 patients found a sensitivity and specificity greater than 70%. Other studies have used segmentation of acne lesions, breaking down the image into several homogeneous regions based on the similarity of their color33.
Given that the severity of acne is the main determinant in the choice of treatment, these evaluation methods based on image processing could help improve therapeutic decision-making. Likewise, their development could suppose a marked potential time saving for the clinician.
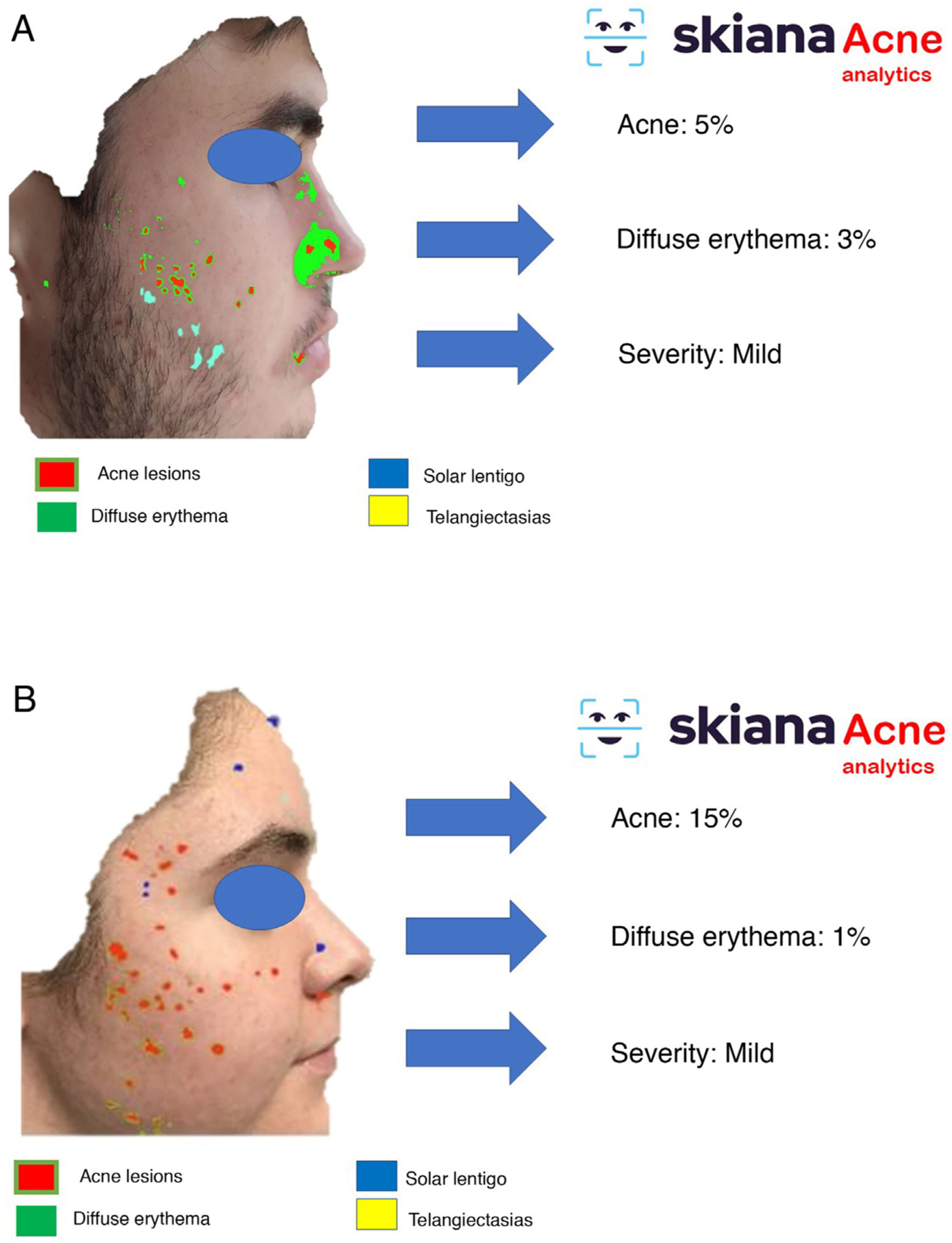
In 2019, Seité et al.34 developed an AI-based smartphone application that graded and classified acne lesions (comedonal, inflammatory, postinflammatory hyperpigmentation, etc.). Apart from the advantages in terms of evaluation of lesions, it was postulated that the use of these applications linked to geolocation could also help assess the impact of external trigger factors such as climate and pollution on the duration and severity of acne.
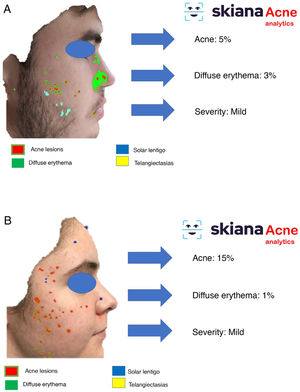
In 2020, Martorell et al.35 developed an AI model for detecting acne lesions in the facial region through a smartphone application SkianaCare®, which calculates the severity for determining whether systemic treatment should complement other treatment, where severity was defined by a cutoff on the global evaluation of acne (GEA) scale (Fig. 6A and B). Thus, in a series of 101 patients, a precision of 97.8% was found in the capacity for definition of degree of severity. In turn, correlation between clinical manifestations and images established the need for systemic treatment, mainly in the form of an oral retinoid, with GEA scores greater than 3 in the facial region.
Other inflammatory diseasesOther AI programs have been designed for diagnosis of different inflammatory skin diseases, such as lichen planus, lichenoid pityriasis, and dermatomyositis from digital images36–39. Huang et al.39 developed a classifier of multiple diseases (based on assessment of 34 attributes such as erythema, scaling, border definition and irregularity) able to differentiate between several skin diseases such as psoriasis, lichen planus, and seborrheic dermatitis.
For the study of dermatomyositis, in addition to assessment of skin images, ultrasound images of muscle have been used to differentiate between normal muscle, dermatomyositis, polymyositis, and inclusion-body myositis38.
Application of artificial intelligence in skin cancersIn the field of skin cancer, of note have been some publications that cover the potential of the new—at the time—information systems.
The first publication, from 1986, talked of how computers could automate diagnosis of facial tumors and issuing of recommendations40. Another from 1992 highlighted the use of computer imaging analyses to aid diagnoses made by professionals with less experience in the field of melanoma41.
Since then, and more notably in the past decade, there has been an exponential increase in publications on AI to support care of patients with skin cancer. This increase can be defined in 2 stages that, although not perfectly differentiated, give an approximate description of the development of the application of AI in skin cancer.
In the first stage, which draws on previous years, there is an emphasis on theory and multiple practical attempts were made. In this stage, we could say that the systems based on the use of AI were honed.
There were publications on the reliability and feasibility of the use of conventional digital macrophotographs to allow the system to distinguish between melanocytic and nonmelanocytic lesions42. It was also observed that the combination of clinical information and imaging helped with diagnosis of basal cell carcinoma43. Many other publications of the same nature highlight that through a combination of different information sources (exhaustive medical history, images obtained from different systems…), greater precision was obtained in automated diagnosis. It was even possible to see how AI could be used to determine surgical margins in intraoperative biopsies44.
Automated prognostic models can also be included in this stage. An example would be computational algorithms that enable prediction of metastatic progression of cutaneous melanoma based on gene expression and microRNA data45.
A second more recent stage has seen AI systems used in (and outside) the clinic with different functions.
One of the most widely disseminated publications, from 2017, proposed using DL algorithms for classification of tumor lesions46. Specifically, the authors compared the degree of diagnostic concordance between the automated system and a group of specialist dermatologists when distinguishing seborrheic keratosis from keratinocyte carcinomas and benign nevi from melanoma, with promising results for use in support of diagnosis. These algorithms could become a tool to extend the reach of dermatologists outside the clinic.
The use of algorithms has been proposed for automated image analysis to support therapeutic decisions (excision versus observation) in the management of pigmented lesions47, or to give initial guidance to the dermatologist who subsequently would make the decision according to clinical context48. Of particular interest is the use of images obtained by free smartphone applications; with these some authors have proposed algorithms for detecting changes in pigmented lesions that could predict progression to melanoma49.
With particular interest in pigmented and melanoma lesions, one of the most prolific groups was that led by the dermatologists in Heidelberg, Germany. In their research, they primarily use CNN-based systems. In the first articles, by Haenssle, preliminary results superior to dermatologists were obtained for recognition of melanoma by dermoscopy in experimental conditions8. The group lead by Brinker has continued to highlight the excellent performance of CNN-based systems for differentiating melanomas from melanocytic nevi by dermoscopy50,51, combining clinical and dermoscopic images52, or with histopathologic images53. In conditions of use closer to clinical practice, and including diagnosis of other types of tumor (not just melanoma), they have achieved results similar to those of dermatologists54.
Other groups have achieved good results in the identification of melanomas from dermoscopic images using DL-based systems55, or acral melanomas using CNN-based systems56.
These systems and approximations can be used in exploring data to generate models predictive of response to immunotherapy in cases of metastatic melanoma57. Some investigators are even using gene expression profiling and microRNA analyses with ML algorithms for molecular distinction between melanocytic nevi and melanoma58.
In the case of nonmelanoma skin cancer, use of AI-based systems appears at present to be supported by less robust evidence59. In any case, CNN-based systems have been successfully used in the automated detection of facial keratinocyte cancers60 and actinic keratosis61 from clinical photographs.
Artificial intelligence in dermatopathologyHistologic diagnosis of skin lesions has a substantial subjective, observer-dependent component. Specially, relatively nonspecific inflammatory lesions, poorly differentiated skin tumors, or diseases with a wide spectrum of presentations that include benign lesions through malignant ones, including borderline cases, can generate substantial diagnostic uncertainty, with differences of opinion even among expert dermatopathologists. Perhaps one of the best examples is histological diagnosis of melanocytic lesions: the difference between dysplastic nevus and in situ melanoma is subtle, and experienced dermatopathologists may sometimes reach different diagnoses for the same lesion2–4.
In dermatopathology, the use of AI as a diagnostic tool started at the end of the past century, with the TEGUMENT project (Fig. 4)62. However, it is only more recently that several studies have started to support (in very preliminary fashion) its potential practical use as an aid for histologic diagnosis.
AI has shown a high precision in performing several essential functions of histologic diagnosis, such as mitosis count and assessment of positivity in immunohistochemical tests63. On the other hand, DL algorithms showed a precision of almost 100% in binary classification of tumor lesions, and almost 80% for classification of these same lesions in 4 categories (basaloid, squamous, melanocytic, and others)63.
With regards the differential diagnosis between nevi and melanoma, CNN-based methods have been assessed in different studies63,64. In 2019, Hekler et al.62 showed overall discordance of 19% between a CNN-based system and an expert dermatopathologist, with this percentage being similar to that between expert dermatopathologists. Also in 2019, Hart et al.65 reported a precision of approximately 90% for binary classification of Spitz nevus and conventional nevus. More recently, in 2021, CNN-based techniques showed a sensitivity, specificity, and precision approaching that of 18 expert dermatologists when assessing 50 melanocytic lesions to discriminate between nevi and melanomas64.
One of the main limitations in the use of CNN-based systems in dermatopathology is their limited classification system: whereas trained dermatopathologists are able to recognize several morphologic variants and perform broad differential diagnoses, current CNN-based models mainly use binary classification systems (they indicate whether an image is positive or not for a diagnosis) and lose precision when more categories are introduced66. In fact, there are hardly any studies in dermatopathology of nontumor lesions, where differential diagnosis is, in many cases, broader and more subjective. Furthermore, given the ground truth in the learning phase of these systems is established by one or several dermatopathologists, and given that many entities lack diagnostic criteria with absolute precision, this fundamental or ground truth is not completely free of subjectivity4.
Therefore, although preliminary results are promising, it is necessary to extend them (to beyond binary systems) and AI systems need to be validated before they can be used in dermatological clinical practice.
Probably, these systems could be used in the future for automating reproducible tasks, such as screening, making them a diagnostic aid to improve not only the daily work of the dermatopathologist, but also an important tool in research and even education.
Artificial intelligence applied to cosmeticsThe field of cosmetic dermatology has been one of the sectors in which multiple models have been developed based on innovative solutions. Currently, on the market, models include virtual planning for cosmetic treatment with neuromodulators and treatment with cosmetic fillers, as well as use of robotics in automation of different laser treatments67.
Home use assistantsThe application of new technologies has generated multiple strategies whose end objective is to empower patients through information and generation of personalized content, with the aim of facilitating decisions on skin and hair care.
Many companies have created intelligent questionnaires about skin and hair care that include questions about patient demographics, skin and hair characteristics, and desired esthetic outcomes to generate personalized recommendations.
Along these lines, the startup known as PROVEN Beauty®, based in California, used ML to offer personalized cosmetic products to consumers for skin care. The algorithm uses information from a large skincare database called Beauty Genome Project, which includes more than 8 million client opinions, more than 100,000 products for skincare products currently on the market, 20,000 ingredients, and more than 4000 peer-reviewed scientific articles on skin and skincare ingredients68.
Augmented reality is another technology that enables personalization of routines for skin and hair care. In these cases, it goes one step further than questionnaire-based models by incorporating patient images in the decision-making process. The company L'Oreal® offers 2 online web-based applications of augmented reality that are free for users. The first application offers online skin analysis through the Vichy® Skin Consult AI® website. This application generates a skincare and antiaging simulation with the capacity to detect, quantify, and predict changes in the skin69. This ‘virtual mirror’ applies computerized vision technology to simulate the result that could be obtained with beauty and cosmetic products. The project was developed with the support of dermatologists to assess skin afflictions such as dyschromia, dryness, and wrinkles. After uploading an image, the system generates information for the consumer on the quality of their skin, aspects to improve, and personalized recommendations for a skincare regimen. Aspects analyzed include infraorbital lines, elasticity, fine generalized lines and deep wrinkles, lack of luminosity, hyperpigmentation, and pores. However, the main limitations of the system include: 1) bias in the recommendations based on a limited set of products; 2) loss of information in the analysis because lateral images are not processed; 3) lack of the possibility for self follow-up; and 4) lack of information generated by the user, and so personalized recommendations that are not only based on the physical appearance of the user cannot be generated.
Likewise, the free smartphone application denoted Skin360®, developed by Neutrogena®, uses the smartphone camera to assess the skin in search of dark marks, baggy eyes, wrinkles, changes in texture, and expression lines. This assessment provides a’ Skin360 score,’ and makes suggestions to the user for skincare products based on these 5 parameters. The patient can also check on improvements with the recommended products over the course of days, weeks, and months, with follow-up assessments using the application. In addition to this score, the application also checks for lifestyle habits that may impact skin health (sleep, exercise, stress, and daily use of the product)70.
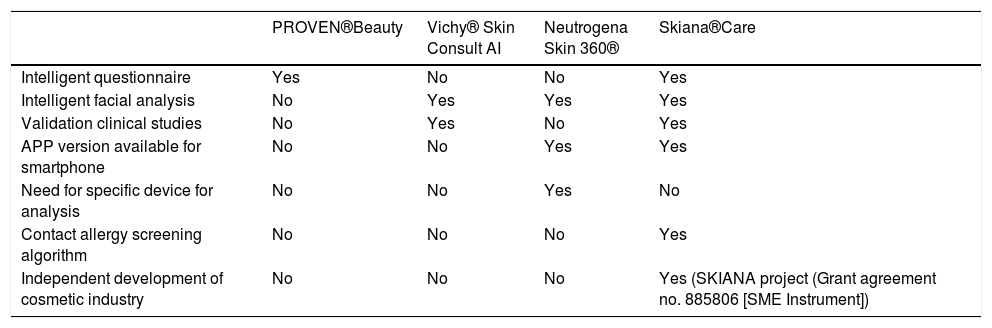
In a similar vein to the above applications, but led by dermatologists, and with the aim of improving a patient’s skincare through cosmetic reinforcement and reducing excess discarding of products by the user due to difficulties in selecting an appropriate product, in 2020, Martorell et al.35 developed the first independent application in the cosmetic industry, with the support of the European Commission within the Skiana Health and AI project® (European Grants SME I 2020, Grant no. 885806) (Table 1).
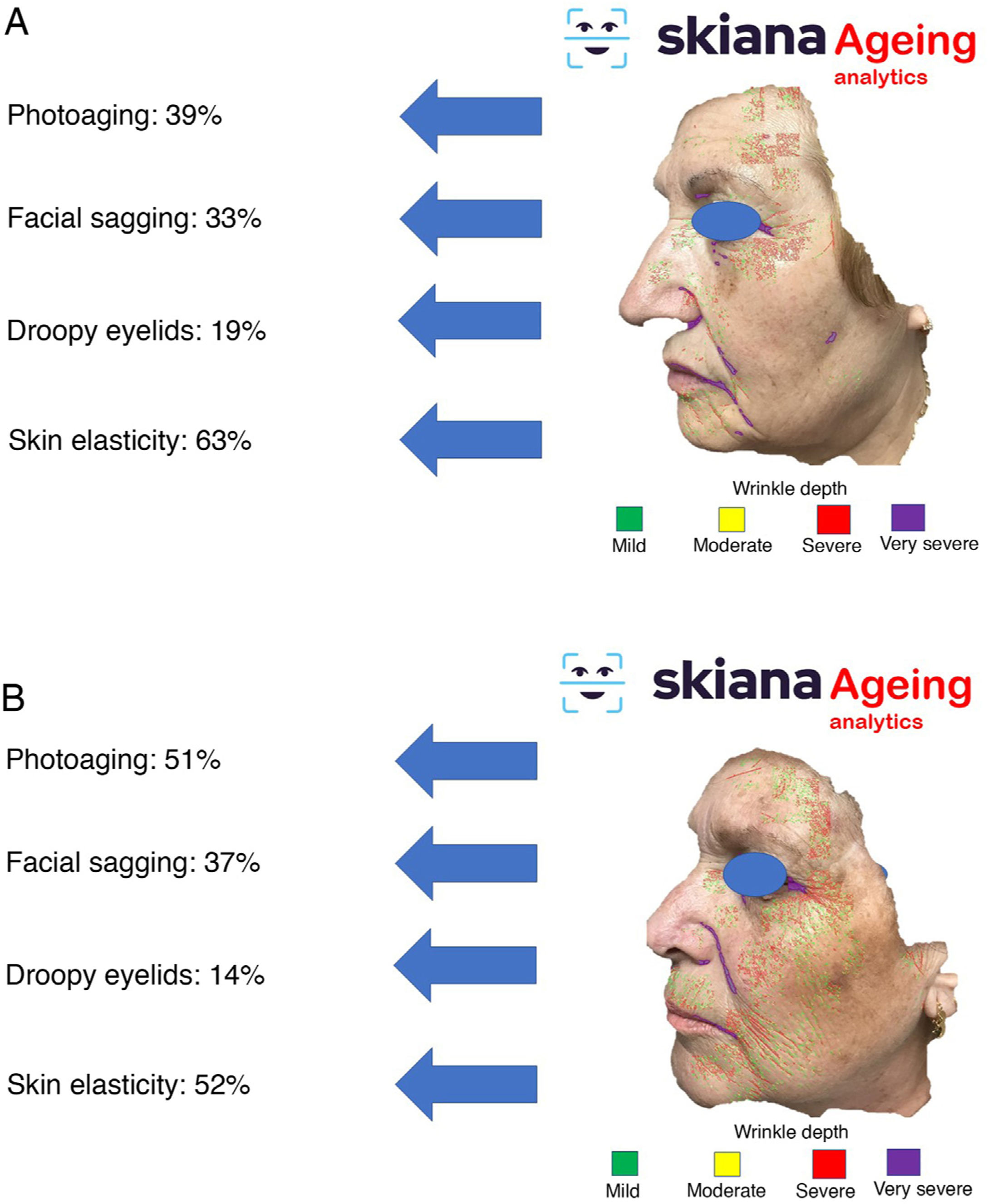
This virtual personal assistant is the first in its generation free of conflict of interest of any pharmaceutical or cosmetic company. It was developed in the cloud and available for Android® in the App Store and Apple® in the Apple App Store under the name Skiana®. The aim, through a combination of a brief intelligent questionnaire and information obtained from a facial analysis generated using AI algorithms, is to improve wellbeing through healthy skin. The main aims are the following: 1) generation of skin cancer prevention campaigns and health education through skin-healthy advice; 2) capture of the end user through generation of objective reports available on their mobile devices that include information on skin health—these data range from quantification of photoaging, flaccidity, elasticity, and changes in skin texture such as quantification of acne lesions (Figs. 6 and 7); 3) generation of patient expert in skincare, providing personalized information on state of skin and training in cosmetics, including information on cosmetic products, in order to help improve skincare; and 4) positioning the dermatology and venereology specialty as a reference in the field of integral skincare.
The main pros and cons of the different virtual assistants are summarized in Table 2.
Main differences between the virtual cosmetic assistants currently available.
| PROVEN®Beauty | Vichy® Skin Consult AI | Neutrogena Skin 360® | Skiana®Care | |
|---|---|---|---|---|
| Intelligent questionnaire | Yes | No | No | Yes |
| Intelligent facial analysis | No | Yes | Yes | Yes |
| Validation clinical studies | No | Yes | No | Yes |
| APP version available for smartphone | No | No | Yes | Yes |
| Need for specific device for analysis | No | No | Yes | No |
| Contact allergy screening algorithm | No | No | No | Yes |
| Independent development of cosmetic industry | No | No | No | Yes (SKIANA project (Grant agreement no. 885806 [SME Instrument]) |
The application of these technologies in the field of robotics, with the development of devices able to produce personalized cosmetic products at home from the information obtained by the applications described above, will be the next step in cosmetics, with companies such as Procter & Gamble® (Opté Precision Skincare System®)71 and L’Oréal® (Perso®)72 being the pioneers in these types of home devices.
Likewise, the use of these algorithms in professional supports that allow images to be taken with different light parameters such as Visia® (Canfield® Inc.) o Lifeviz® micro (Quantificare® Inc.) will help the cosmetics professionals to improve the skincare of their patients in their clinics73.
Models to predict response by artificial intelligenceThe application of the DL model and generation of neural networks will help increase the effectiveness and safety of cosmetic procedures.
Examples along these lines include the application of DL to best define the cosmetic treatment option according to facial wrinkle depth74–76. The potential for DL to predict clinical response to laser therapy has also been evaluated. For example, Cazzaniga et al.77 applied this technology to the prediction of duration of excimer laser treatment for patients with vitiligo, with promising results. With data from their clinical trial, the model achieved an overall precision of 66.46% in predicting the time to repigmentation in responders. Given that treatment can be slow and costly, a predictive model for clinical response and duration of treatment would help both patients and healthcare providers decide whether to continue with treatment. The models of clinical outcome can also be applied to other types of laser and treatment modalities for treating afflictions such as acne scars, dyschromia, and vascular disorders to help establish realistic expectations in interested patients.
Artificial intelligence in skin radiology and ultrasoundAmong the radiological imaging techniques, both simple computed tomography (CT) and magnetic resonance (MR) imaging are used essentially in the context of staging or follow-up of melanoma. The advantage of these techniques is that they can be readily standardized, and there are large image databases for training the algorithms.
Techniques based on ionizing radiation, such as CT in dermatology patients, have been used for detecting melanoma metastasis. In a study by Aissa et al.78, using ML techniques and vessel suppression, algorithms were able to find additional metastases in 54.3% of cases. However, authors concluded that diagnosis of additional metastases would not have affected patient management.
Regarding the use of MR, AI has been applied to the detection and diagnosis of lesions. Kniepp et al.79 reviewed 189 brain metastases, including from melanoma, and using decision tree algorithms in combination with clinical data, they demonstrated greater capacity to differentiate between the metastases of other tumors (squamous cell carcinoma, microcytic lung cancer) than radiologists, with an area under curve of 0.82. This use of radiologic imaging to infer histological diagnosis of lesions has been denoted as radiomics, and is one step beyond image detection or segmentation.
Skin ultrasound is a technique that offers advantages over other imaging techniques as it is harmless, fast, and available at the patient’s bedside79. However, ultrasound has several limitations as source of data for AI, the main ones being the absence of large ultrasound imaging banks suitable for training and validating algorithms and the intrinsic characteristics of ultrasound scans, which means they are not as standardized as is the case with CT and MR images.
Nevertheless, in recent years, the development of AI in ultrasound has started to provide evidence regarding how it could be applied in the 3 main ultrasound issues, and specifically, in the case of skin ultrasound: classification, regression, and segmentation80.
ClassificationMost classification algorithms have been applied to ultrasound diagnosis of breast or hepatic lesions81,82. Although there are numerous publications that have attempted to reduce the key features needed for AI-assisted diagnosis83, the number of images available is usually less than 300, and they have been captured in different clinics with different validation protocols, and so the results are not usually comparable84.
From the commercial point of view, some algorithms have been developed for assisted diagnosis, mainly using breast and thyroid ultrasound85–87. The algorithm suggests a risk of malignancy of the lesion selected by the ultrasonographer, but it does not indicate a specific diagnosis for the lesion studied. However, these algorithms have not been validated by any regulatory agency.
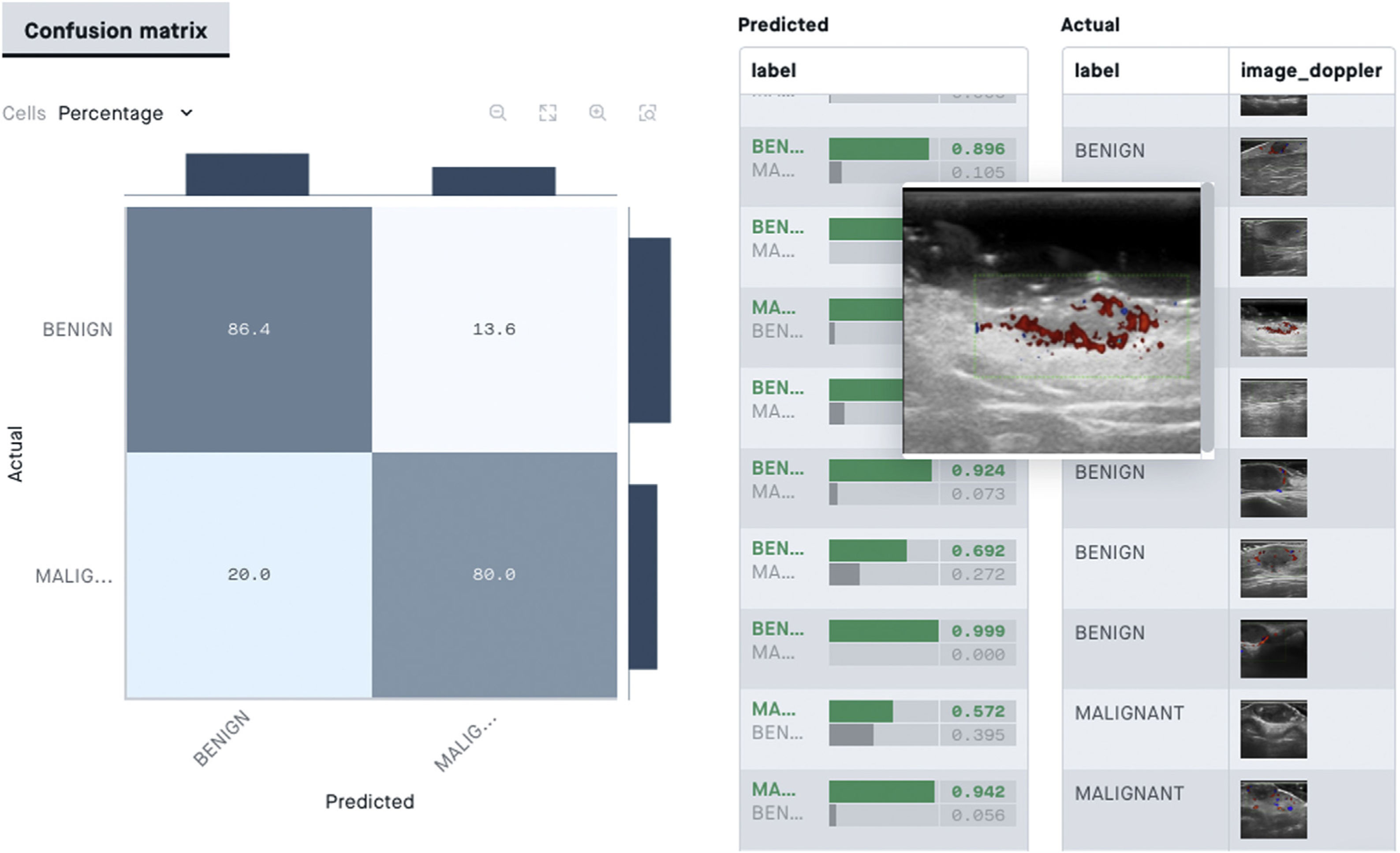
From the point of view of skin ultrasound, classification algorithms could be useful in AI-assisted diagnosis of skin lesions. In a recent study by Alfageme et al.88, a DL-based algorithm with predetermined architecture, trained with a bank of 235 ultrasound images of benign and malignant skin lesions, was able to correctly classify the lesions at a similar rate to an expert human evaluator (77.1% versus 74.1%) who was exposed to the same image bank.
In the case of melanocytic lesions, using ultra-high frequency ultrasound devices (70 MHz probes), the group led by Romanelli also reported a capacity to differentiate between melanocytic nevus with an accuracy of 76.9% and a sensitivity and specificity for diagnosis of melanoma of 80% and 74%, respectively by applying DL-based algorithms89.
With skin ultrasound, it remains to be seen what the optimum characteristics are for appropriate image banks for this application in terms of equipment, resolution, and similarity with inflammatory lesions, which may hinder the applicability of these algorithms in clinical settings.
RegressionRegression involves estimating continuous values instead of discrete classes of data. DL has been applied to regression, for example, for estimating the orientation of muscle fibers from ultrasound images or gestational age.90,91 With regards skin ultrasound, regression algorithms could be useful for assessing the duration of skin lesions, automatic determination of ultrasound Breslow depth, or in algorithms applied to the study of skin ageing and its treatment92.
SegmentationSegmentation is the ultrasound delimitation of the structural limits of a lesion. Automated ultrasound segmentation is a challenge, as ultrasound images are affected by scintillation, shadows, and absence of limits, as well as frequency compensations, depth, and ultrasound resolution during image acquisition93.
Several ultrasound segmentation approaches have been developed, including methods based on intensity thresholds, sets of levels, and active contours94. The approaches based on intensity are sensitive to noise and image quality95. Active contours and sets of levels require an initialization that may impact the results. Most of the conventional approaches are not totally automated.
ML-based segmentation methods usually involve 2 steps: first, a pixel classification of the target structure, followed by a cleaning or softening step, as pixel classification is noisy from the ultrasound point of view. In recent articles, several classification approaches have been investigated with specific focus on the application of several types of neural networks, including DL96.
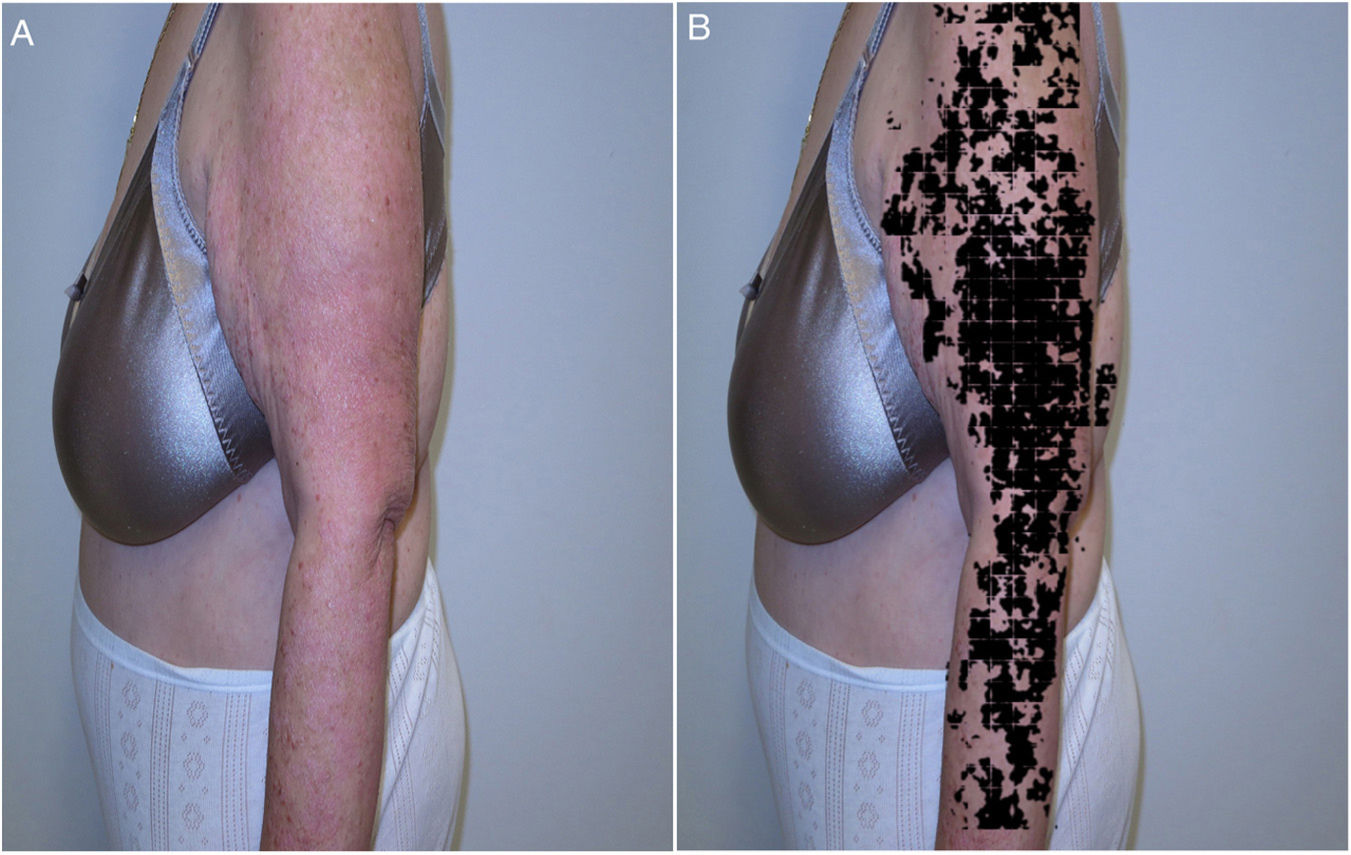
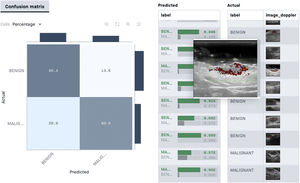
In skin ultrasound, the segmentation algorithms could be useful for detecting lymph node metastasis or for ultrasound delimitation of skin lesions (Fig. 8).
Ethical and regulatory aspects of AI in dermatologyWe may not fully realize how unique the situation we are facing is, as never before in human history have we had the capacity to compile such large volumes of data.
The internet of things and big data can feed the AI and ML algorithms. The more complex these algorithms, the greater their predictive power, and so all efforts are focused on correlating a vast number of variables to achieve accurate predictions. The undeniable aim is to achieve Laplace’s demon, a machine able to predict any scenario in a fully deterministic fashion97. For this reason, AI is also being studied within the field of bioethics.
Of note is that the European Parliament, on proposing a Letter on Robotics, started listing bioethical principles as the rules to be followed for robotics engineers, and compiling the ethical principles that should guide AI, which have been denoted “Asilomar Principles for Artificial Intelligence”98.
In any case, the ethical framework for AI in dermatological practice should also reflect the basic principles of medical ethics: autonomy, benefit, absence of harm, and fairness. But in the specific case of AI, this ethical framework should also include specific sections, such as transparency and accountability99.
The principle of autonomy refers to the fact that the patient has the right to make his or her own decisions. In medical care, informed consent is the mechanism by which autonomy is ensured, but dermatology images not only include information in the form of pixels but also information on health (at least when the image is linked to a patient’s medical history), such as demographic data and institutional information. When these images are used for training AI algorithms, informed consents should be adapted for this purpose, such that these images can continue to be used.
The principles of benefit and absence of harm are 2 sides of the same coin and are obvious in conventional medical care, but certain contradictions may be present in the case of AI, as the algorithms may benefit from stratification with many parameters, improving diagnostic yield and, therefore, improving the patients’ wellbeing, but at the same time, such segmentation could be used for profit.
The principle of fairness requires a fair distribution of medical products and services. The development of AI should encourage equity, removing certain discriminations and preventing certain implicit biases, as will be discussed later.
Finally, the principles of transparency and accountability may be addressed by the principle of explainability. If an AI system fails or causes damage, we should be in a position to determine the underlying reasons and it should be possible to perform an audit of the entire process. This latter point is surely the main ethical limitation of AI, as these systems are considered a type of black box in which it may be possible to understand the logical processes and mathematics behind the algorithm but not the downstream data processing, such that the result is, in reality, unpredictable100.
This all ties in with the debate about accountability, when the AI systems automatically trigger diagnostic or therapeutic decisions, we may well ask who is accountable if the system fails101. The question can be extrapolated to other fields, and this debate came to the fore in March 2018 with the first fatal accident with a self-driving car in Tempe, Arizona, United States. Was the person who was in the car responsible? Or the car manufacturer? Or the software developer? There is no simple answer. Above all, it is necessary to develop regulations and legislation on the topic to ensure there are no legal grey areas. When AI is applied in medicine, these considerations are even more important102.
Another important question is whether algorithms may perpetuate human bias precisely because they work like a black box. When the algorithm is trained with biased data, because underrepresented populations are not included, existing inequalities may be reproduced for further entrenched103. If the data acquired by the algorithm are not sufficiently inclusive and balanced, the system could learn to take unfair decisions (this is very obvious in facial recognition systems, which are more reliable with white males than black women)104.
In dermatology, although there are many AI applications, in the initial phases, the focus has been on diagnosis of pigmented lesions, above all, melanoma. The reality is that, for the time being, most AI and ML programs are learning mainly from images of people with light phototypes. For example, in the International Skin Imaging Collaboration: Melanoma Project, one of the most widely used and with open access code, most of the patients have light phototypes and come from populations in the United States, Europe, and Australia. As a result, the algorithm is not very reliable for lesions on black skin. Paradoxically, melanoma is usually detected in more advanced phases with worse survival rates in Afro-American patients. The algorithm only improves early detection in light skin types, with no benefit for patients with black skin, thus clearly worsening the biases already observed in society104. The solution is not easy to implement but it is simple: effort has to be made to train the algorithm in all skin types.
From the regulatory point of view, several organizations have published frameworks for setting up AI applications in the field of medicine. This is all rather complex, starting from the fact that although legislation requires the absence of ambiguity, there is currently no clear definition even for what we mean by the term artificial intelligence.
The second crucial point for the legislators is whether to consider the AI software used in medicine as a medical device (there is a distinction between programs that analyze data to have a deeper knowledge of a disease than those that make diagnostic or therapeutic decisions in individual patients). On this point, we can differentiate between the European vision and that of the United States105.
In Europe the legislation in force is the Regulation on medical devices, which was signed in May 2020 and which replaces Directive 93/42/EEC106. As this is a regulation (rather than a directive), it applies directly to territories in the European Union without the need to be implemented by the different member states. This regulation requires that manufacturers ensure that the devices they produce meet a series of essential requirements that will depend on the potential risks of each and that, at times, will need certification by an independent body.
In the United States, the regulatory body is the US Food and Drug Administration since the end of 2016, when jurisdiction over software used in the health field was transferred to this body by the 21st Century Cures Act107, with a more restrictive reading in terms of introduction of unsupervised diagnostic software.
In addition, all these tools that feed off a huge wealth of data have other implications associated with privacy and data protection, as well as cybersecurity. As a result of these potential threats, the European Union decided to update the legislation, and the General Data Protection Regulation has been in force since May 24, 2018, replacing the previous legislation. The new legislation requires user consent, and the user should be aware of the purposes, with marketing of these data by third parties being explicitly excluded108. In the United States, computer-related security in the healthcare field is covered by the Health Insurance Portability and Accountability Act.
In 2018, the European Commission established the High Level Experts Group on Artificial Intelligence, with the aim of supporting the implementation of the European strategy for AI, including drawing up recommendations for future development of norms that regulate ethical, legal, and social aspects of AI.
According to these fundamental rights and ethical principles, these guidelines have 7 key requirements that any AI system should meet to be reliable: 1) It should not impact human autonomy; 2) it should be technically sound and safe; 3) privacy and data management should be respected; 4) it should be transparent; 5) it should be diverse, with no discrimination or inequality; 6) it should respect the environment; and 7) there must be accountability109.
AI will play an important part in the coming years in our specialty, and we need to be guided by ethical principles, such as the principles of benefit and respect towards our patients. We should remember that a dermatologist is not a simple interpreter of images, and that our functions also include many other aspects that cannot be performed by machines.
ConclusionsWe are living and will live a full-blown AI revolution in dermatology, with a range of systems of differing focus and undoubted potential.
A recent literature review claims that neural-network based AI systems (whether in their deep or convolutional modalities) have a greater capacity for recognition than dermatologists110. While this affirmation is open to debate and requires context, it is nevertheless true that AI systems are progressively improving.
On the one hand, conventional automated image analysis systems have been easily surpassed by neural network systems used in the past 10 years111.
On the other, the systems are now capable of processing a huge additional amount of information to achieve a reasoning analogous to that of a dermatologist (for example, comparing with other pigmented lesions of the patient to avoid false suspected diagnoses in the context of a predominance of atypical nevi)112.
After analyzing the opinion of different authors, we believe that progress in these technologies needs to be progressive, safe, and based on the generosity of the dermatology community, as publicly available databases with images and broad clinical information are essential to improve the reliability of these systems113. These systems could undoubtedly be a support for other physicians if no dermatologist is available and cannot be reached on short notice. Nevertheless, we believe that the future will not see AI systems replacing dermatologists, but rather they represent an opportunity for improving clinical practice thanks to the many advantages that these systems may offer.
In short, the optimal situation would be a symbiosis between AI and dermatologists to improve decision-making for our patients, conscious that the ideal is a combination of human intelligence with AI114, maintaining a fluid relationship between physician and patient32. It is therefore essential to change the current tendency; dermatology specialists should take the initiative in the development and orientation of the AI products focused on covering unmet needs in current medical practice.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Dr. Antonio Martorell has received fees for participation in advisory boards and satellite symposia with the following companies: AbbVie, Amgen, Janssen, UCB, Lilly, Novartis, LEO Pharma, Sandoz, and MSD.
Please cite this article as: Martorell A, Martin-Gorgojo A, Ríos-Viñuela E, Rueda-Carnero JM, Alfageme F, Taberner R. Inteligencia artificial en dermatología: ¿amenaza u oportunidad? Actas Dermosifiliogr. 2022;113:30–46.